Figure 7.
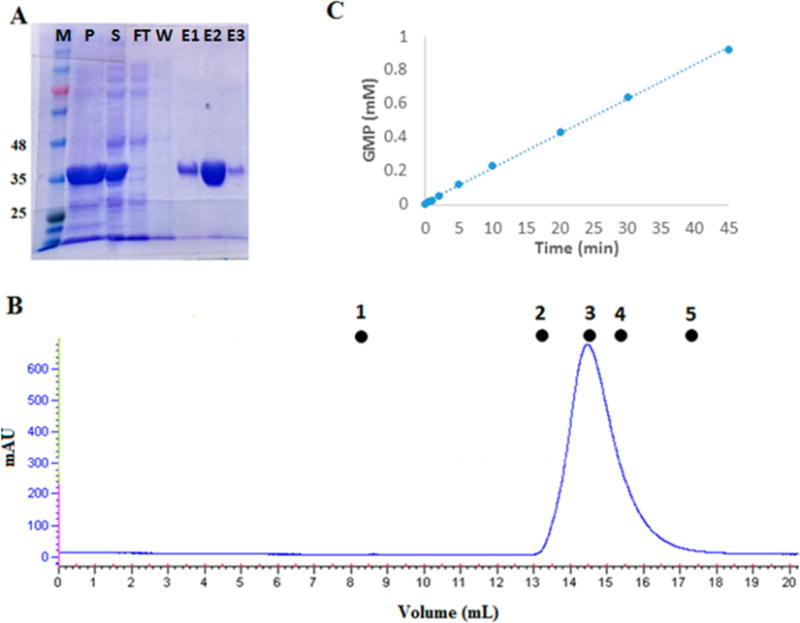
Phosphodiesterase activity of the isolated PDE domain purified from transformed E. coli T7 Express cells. (A) Coomassie-stained SDS–PAGE gel showing fractions from purification of the isolated PDE domain from transformed E. coli T7 Express cells using a six-His tag with a Ni affinity column: lane M, molecular mass markers in kilodaltons; lane P, pellet from the cell extract; lane S, soluble fraction from the cell extract; lane FT, flow-through, nonbound fraction from the Ni affinity column; lane W, last wash before elution with an imidazole gradient; and lanes E1–E3, three representative fractions from the imidazole gradient eluate. (B) AKTA FPLC profile for size exclusion chromatography of the isolated PDE domain on a Superdex-200 10/300 GL column. Molecular mass standards: (1) blue dextran, 2000 kDa, 8.5 mL (void volume); (2) aldolase, 158 kDa, 13.15 mL; (3) albumin, 67 kDa, 14.57 mL; (4) ovalbumin, 43 kDa, 15.41 mL; and (5) chymotrypsinogen, 25 kDa, 17.35 mL. The isolated PDE domain elutes at 14.5 mL, giving a mass of 74 kDa, consistent with a homodimer. (C) HPLC assay of the cGMP phosphodiesterase activity of the isolated PDE domain. The concentration of PDE in this assay was 100 nM, which gives an apparent kcat of 3.13 ± 0.12 s−1.
