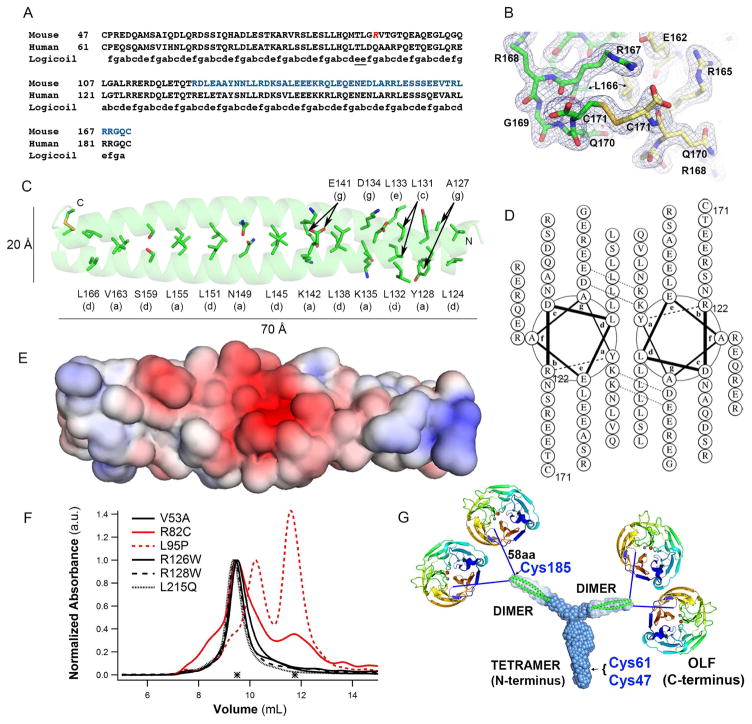
Figure 4. Crystal structure of mLZ122-171, overall myocilin model, and effects of selected glaucoma-relevant variants.
(A) Partial sequence alignment of human and mouse myocilin with predicted register of heptad repeats. Red Arg, start of the construct used for crystallization (mLZ93-171). Blue stretch, crystallized fragment mLZ122-171. (B) Two mLZ122-171 helices are covalently tethered by a C-terminal disulfide bond. Final 2Fo-Fc (blue) electron density map is contoured at 1.0 σ and Fo-Fc (red/green) is contoured at ±3.5 σ. Chains A and B are labeled green and yellow. Stereo image presented in Figure S5A. (C) Ribbon diagram of mLZ122-171 parallel dimer with labeled heptad positions (a) and (d) and disulfide represented as sticks. (D) mLZ122-171 in helical wheel representation. (E) Electrostatic surface (negative (red, -5 kT/e-) to positive (blue, + 5 kT/e-)) of mLZ122-171 dimer. (F) SEC profile comparison of six NTD33-226 glaucoma-relevant variants reveals altered elution and thus quaternary structure for R82C and L95P (red traces). (G) Model of full-length myocilin: Y-shaped dimer-of-dimers composed of an N-terminal parallel tetrameric domain leading to two parallel CC dimers, followed by 58 amino acid long linkers, and C-terminal OLF domains. Cys185 is located just prior to the start of the 58aa linker; Cys47 and Cys 61 are within the N-terminal tetramer but locations within the stem are not precisely identified. See also Figure S1, S5.