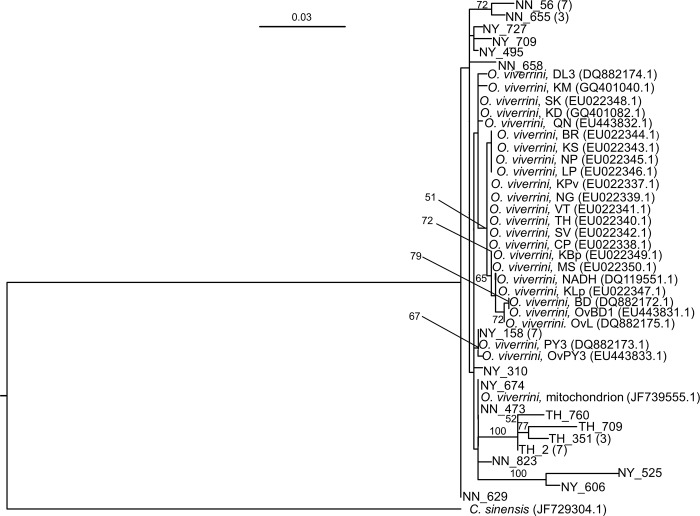
Fig 5. The Randomized Axelerated Maximum Likelihood tree based on the nad1 sequences of O. viverrini.
The alignment of 688 nucleotide sequences without gaps and 18 variants from three villages (NY = Na-Yao, NN = Na-Ngam, TH = Thoong-Heang) were analyzed. O. viverrini Thai isolates (BR, KS, LP, NP, KPv, MS, KLp, KBp, SK, NADH1), Lao PDR isolates (CP, NG, SV, VT, KM, O. viverrini mitochondrion partial genome), Vietnamese isolates (BD, OvBD1, OvL, PY3, OvPY3, QN, DL3) and a Cambodian isolate (KD) were used as the reference isolates. The percentages of 1,000 replications (bootstraps) of more than 50% are shown at the nodes. The number of samples is in parentheses.