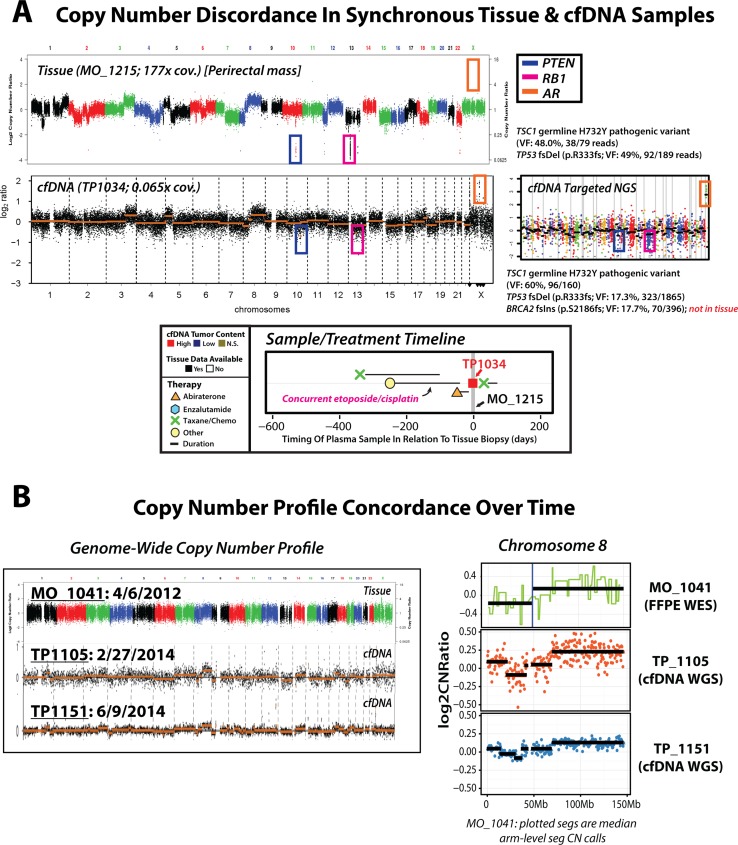
Figure 4. Unique precision oncology considerations identified via serial and synchronous tissue and cfDNA NGS-based profiling in patients with advanced prostate cancer.
(A) Genome-wide (tissue and cfDNA) and targeted (cfDNA only) NGS copy number profiles are displayed, along with treatment and sample timeline, for synchronous (same-day) tissue and cfDNA specimens from a patient with metastatic castration-resistant prostate cancer (mCRPC) with a history of both primary prostatic adenocarcinoma and a metastatic lesion with small cell carcinoma/neuroendocrine features. Tissue whole exome sequencing (WES) copy number analysis of a frozen perirectal mass tissue biopsy specimen (top left) revealed focal, deep deletions in both PTEN and RB1, and no AR copy-number alterations, consistent with histological reports of high-grade poorly differentiated carcinoma with neuroendocrine features. Individual dots in tissue WES copy-number profile represent exon-level copy-number estimates displayed in genome order, and dots are colored by corresponding chromosomes. cfDNA low-pass whole genome sequencing (WGS) copy number profiling identified focal deep deletions in PTEN and RB1, as well as high level focal AR amplification, highlighting circulating evidence of both AR-driven and AR-independent clones. Individual dots in cfDNA low-pass WGS plot represent bin-level copy-number estimates displayed in genome order (left to right), with segmented copy-number alterations represented by orange horizontal lines. Both tissue (WES) and cfDNA (targeted NGS; copy-number prfile ) mutation profiling identified a TSC1 germline H732Y pathogenic variant (tissue: 48% variant fraction (VF), 79 covering reads; cfDNA: 60% VF, 160 covering reads) and somatic TP53 frameshift deletion (p.R333fs; tissue: 49% VF, 189 covering reads; cfDNA: 17%, 1865 covering reads), while cfDNA targeted NGS identified a BRCA2 frameshift insertion (p.S2186fs; 18% VF, 396 covering reads) not present in the tissue sample, further supporting detection of multiple clones via cfDNA PRINCe assessment. The cfDNA targeted NGS copy number profile is presented at right, showing confirmation of focal PTEN and RB1 deletions along with high-level focal AR amplification as seen by low-pass WGS. Zoomed view of treatment and sample timeline for this patient is presented at bottom, as previously described (see Figure 3). (B) Genome-wide (left) and chromosome 8 (right) copy number profiles from multiple biospecimens taken over time from a single patient with metastatic castration-resistant prosate cancer (mCRPC) (tissue id: MO_1041; cfDNA ids: TP1105 and TP1151). WES of a formalin fixed paraffin embedded (FFPE) tissue biopsy specimen (top left) revealed low but detectable tumor content, and identified copy-number loss affecting 8p and arm-level gain of 8q (at right). Low-pass WGS of a cfDNA specimen collected almost 2 years after tissue biopsy (TP1105, middle left) revealed elevated cfDNA tumor content with frequent copy-number alterations genome-wide, including copy-number loss affecting chr8p and arm-level gain of chr8q (displayed at right), as detected in initial tissue profiling. A subsequent cfDNA sample (TP1151) again showed detection of elevated cfDNA tumor content and a highly concordant genome-wide copy number profile, with faithful representation of the 8p loss and 8q gain events detected in previous specimens. Overall, these results highlight the consistent representation of early genomic events as inferred from circulating tumor DNA profiled in our cohort.