Abstract
The nuclear pore complex controls the passage of molecules via hydrophobic phenylalanine-glycine (FG) domains on nucleoporins. Such FG domains consist of repeating units of FxFG, FG, or GLFG sequences, many of which are interspersed with highly charged amino acid sequences. Despite the high density of charge in certain FG domains, if and how charge influences FG-domain self-assembly and selective binding of nuclear transport receptors is largely unexplored. Using rationally designed short peptide sequences, we determined that the charge type and identity of amino acids surrounding FG sequences impact the structure and selectivity of FG-based gels. Moreover, we showed that spatial localization of the charged amino acids with respect to the FG sequence determines the degree to which charge influences hydrophobic interactions. Taken together, our study highlights that charge type and placement of amino acids regulate FG-sequence function and are important considerations when studying the mechanism of nuclear pore complex transport in vivo.
Introduction
The nuclear pore complex (NPC) is a megadalton structure that controls the exchange of material between the nucleus and cytoplasm (1, 2, 3) through a combination of passive and facilitated diffusion. Above an ∼30-kDa cutoff, proteins require complexation with nuclear transport receptors (NTRs) to efficiently translocate at rates of nearly 1000 molecules/s (4, 5, 6, 7). This fast translocation rate relies on transient interactions between hydrophobic phenylalanine-glycine (FG) domains on intrinsically disordered FG-containing nucleoporins (FG nucleoporins) and hydrophobic patches on NTRs (1, 5, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20). Without this assistance from NTRs, proteins largely remain excluded from the NPC. Despite the necessity of FG domains (which contain repeating units of FG sequences such as FxFG or GLFG) for facilitated diffusion and self-assembly of the selective matrix, how hydrophobic FG domains exhibit such selectivity for specific hydrophobic domains on NTRs and what parameters tune the molecular recognition necessary for facilitated transport remain open questions. Sequence analysis and MD simulations predict that the biochemistry and charge surrounding individual FG sequences should play an essential role in FG-mediated molecular recognition and therefore determine the organization and selectivity of the NPC (4, 6, 21, 22, 23, 24). However, due to the complexity and redundancy of FG nucleoporins within NPCs, systematic biochemical dissection of the amino acid space surrounding individual FG sequences has remained an experimental challenge.
Here, we investigated whether and how electrostatic interactions surrounding FG sequences tailor both the self-assembly of FG sequences and the selective recognition of hydrophobic substrates. For systematic dissection of how charge type and localization influence FG-mediated selectivity, full FG domains are too degenerate and complex to provide insight into how single amino acids contribute to selectivity. In peptide research, rationally designed peptides and polypeptides have previously been helpful to identify individual amino acids or short peptide sequences important to the function of much larger polymer-based biomaterials such as extracellular matrices (25), silk proteins (26, 27), and elastinlike polypeptides (28, 29, 30). Here, we applied this approach and created systematically varied peptide sequences to identify how amino acids surrounding FG domains in native nucleoporins may contribute to selectivity. Although peptides may not recapitulate all properties of the original protein, they are a suitable model system for this study because they yield insight into the contributions of individual amino acids and their positioning to overall protein function, a task not feasible with intact proteins. Our data demonstrate that the identity and charge of amino acids surrounding FG sequences can impact the structure and stiffness of FG-based gels. Moreover, we determined that charged amino acids enable FG-containing peptide gels to discriminate substrates of varying charge and hydrophobicity. Last, we found that the distance at which a charged amino acid is localized with respect to the FG sequence determines the degree to which charge influences hydrophobic interactions. Together, our data suggest that FG domain function may be determined by the placement and type of amino acids surrounding FG sequences.
Materials and Methods
Sequence analysis
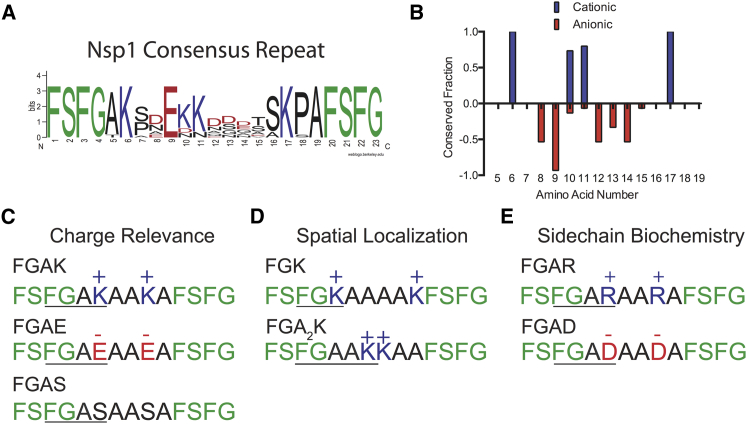
The logo of the consensus sequence was generated using Berkeley’s WebLogo software (31) by aligning 15 repeats according to FSFG as reference. Conserved fraction was calculated for positive charges (number of cationic residues K or R)/(15 repeats) and similarly for negative charges (number of anionic residue E or D)/(15 repeats) at each position indicated.
NTF2 expression and labeling
pQE30-NTF2-6xHis was a gift from the Görlich lab and transformed into DH5α cells for cloning. A cysteine (C) was inserted for fluorescent labeling after the 6× His-tag using standard site-directed mutagenesis with the two primers (5′-CTCAGCTAATTAAGCTTAGCAGTGATGGTGATGGTGATGAGATCTG-3′ and 5′-CAGATCTCATCACCATCACCATCACTGCTAAGCTTAATTAGCTGAG-3′) to form pQE30-NTF2-6xHis-C. The W7A-C mutant containing the terminal cysteine was created by using pQE30-NTF2-6xHis-C and applying standard site-directed mutagenesis with primers 5′-TGAGGAGCCAATTTGTTCCGCGATCGGTTTATCACCCATG-3′ and 5′-CATGGGTGATAAACCGATCGCGGAACAAATTGGCTCCTCA-3′. Expression of NTF2-C and W7A was completed in OverExpress C41(DE3) cells (Lucigen, Middleton, WI) and purified using standard nickel column purification and ion-exchange columns. Labeling of NTF2-C and W7A-C was completed using Fluorescein-5-maleimide (catalog No. F150; Thermo Fisher Scientific, Waltham, MA) in accordance with the manufacturer’s protocol; labeled product was gel purified. Labeling efficiency was ∼50% (data not shown). The final concentrations of NTF2 and W7A for transport were adjusted to 10 μM with 10% of the population labeled.
Peptide and gel preparation
Unless specified otherwise, all chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Peptides were prepared by the Koch Institute Biopolymers and Proteomics Facility (Massachusetts Institute of Technology, Cambridge, MA) and Boston Open Labs (Cambridge, MA). All peptides were HPLC-purified unless specified otherwise, desalted using reverse phase HPLC with 0.05% trifluoroacetic acid (TFA), and lyophilized after synthesis with >95% purity. For fluorescently labeled peptides, a 5-carboxyfluorescein (Anaspec, Fremont, CA) fluorophore was added to the N-terminus, and the C terminus was modified to be an amide. The dye labeling protocol was as follows: peptide-resin that contained an N-terminal free amine but was otherwise fully protected was washed six times with dimethylformamide (DMF) followed by six washes with dichloromethane (DCM) and dried. The material was subsequently reconstituted with dry DMF to reswell the resin. The dye-labeling cocktail consisted of a 4× molar excess over peptide resin of 5-carboxyfluorescein, N,N′-diisopropylcarbodiimide (DCM), and hydroxybenzotriazole reconstituted in a minimal volume of dry DMF and stirred overnight at room temperature in the dark. The cocktail was allowed to preactivate for 30 min before being added to the peptide-resin. Dye-labeled peptide resin was washed six times with DMF, then six times with DCM, and dried in preparation for standard luorenylmethyloxycarbonyl chloride cleavage and deprotection. All quality-control analyses for purity were provided by the Massachusetts Institute of Technology’s Proteomics facility or by Boston Open Labs (Data S1 in the Supporting Material). Neutral hydrophilic (n) and hydrophobic (n) fluorescent reporters could not be HPLC-purified due to aggregation and were used as crude samples.
Fluorescent peptides were diluted into 200 mM NaCl with 20 mM HEPES [pH 7] at 10 μM final concentration for diffusion experiments. Gel peptides were all dissolved in 20 mM NaCl, 20 mM HEPES [pH 7] at 2% (w/v). To facilitate solubilization and gel formation, peptides were vortexed for 30 s and briefly sonicated in a model No. 2510 Bath Sonicator (Branson Ultrasonics, Danbury, CT) to reduce aggregation.
Capillary diffusion assay and analysis
Borosilicate square capillaries (1.5 inch) with 9-mm cross-sectional width (Catalog No. 8290; Vitrocom, Mountain Lakes, NJ) were loaded by piercing premade hydrogels. Ten-micromolar solutions (200 mM NaCl, 20 mM HEPES [pH 7]) of fluorescent peptides were injected into the capillary and sealed with a 1:1:1 (by weight) mixture of petroleum jelly, lanolin, and paraffin. Time lapses of peptide diffusion were taken at 1-min intervals for up to 5 h on a Ti Eclipse inverted microscope using either a CFI Plan UW 2× or an AxioObserver D.1 with an EC Plan-Neofluar 1.25×/0.03 WD = 3.9 (Nikon, Melville, NY) and a model No. C11440-22CU camera (Hamamatsu, Hamamatsu City, Japan). All fluorescence profiles were obtained by averaging the fluorescence intensities across the width of the capillary in the software MATLAB (The MathWorks, Natick, MA). Normalized concentration profiles were obtained by normalizing fluorescence intensities to the bath concentration of the capillary at the initial time point. The fluorescence signal was linear up to 50 μM (Fig. S2) when testing FAM alone in FGAK gels. To plot concentration profiles, the signal was not adjusted past the saturation point; data therefore represent the lower bound of the actual concentration of reporters accumulating in the gel. All data represent at least three independent replicates. Student’s t-test was applied to determine p-values between experimental conditions.
Effective diffusion rates were fit by minimizing the squared error of a simulated concentration time course in a region of the capillary on the gel side of the interface over a 100-min window. To achieve this fit, we numerically solved the diffusion equation for the concentration of probe c:
using MATLAB’s pdepe function (The MathWorks). The initial condition c(x,0) was set by the concentration profile at the first timepoint, and the boundary conditions c(0,t) and c(L,t) (for a fit over length L) were similarly determined by the concentration profiles at the edges of the region of interest. The D that minimized the squared difference between the simulated and actual concentration profiles was the value reported; minimization of error took place iteratively using a modified gradient descent algorithm. The window from 150 to 250 min was generally used for fitting, but for particularly fast-diffusing probes (D > 1000 μm2/min), windows of 50–150 or 30–130 min were used. The earlier time period allowed for the fitting to take place before steady state or pseudo-steady state was reached, which was crucial for precise fitting. Fitting was designed to take place away from the interface to avoid interfacial effects, and the interface was determined in one of two ways. If there was an interfacial peak, fitting began at the x value to the right of the peak where the concentration reached 0.5 times the maximum concentration. If there was no interfacial peak (generally with a hydrophilic probe and a charge-matched gel), the interface was identified as the main inflection point of the initial concentration profile, because the buffer was initially loaded with peptide and the gel was not. The inflection point was identified as the minimum (most negative value) of the numerical first derivative. Fitting was started 20 pixels to the right of the interface. Each fit region was also manually inspected to confirm that it was located at the approximately correct location. Examples of the diffusion-coefficient analysis across the time series as well as the general fit appear in Fig. S3.
Nile Red fluorescence assay
Using the diffusion methods mentioned above, Nile Red dye (N1142; Thermo Fisher Scientific) was loaded into capillaries at 10 μg/mL in 200 mM NaCl, 20 mM HEPES [pH 7].
Rheological testing
Rheological tests were performed on a model No. MCR 302 Rheometer (Anton Paar, Graz, Austria) in a cone-plate geometry with a 25-mm diameter, 1° cone angle, and 51-μm truncation. The temperature was maintained at 25°C and evaporation was controlled with an H2O-filled solvent trap. To identify the linear regime, amplitude sweeps were conducted at ω = 10 rad/s from γ0 = 0.01–100% strain. In the linear regime, frequency sweeps were conducted using the previously determined strain amplitude from ω = 100 rad/s to 0.1 rad/s.
Transmission electron microscopy
Images were taken with a JEOL-1200 transmission electron microscope (JEOL, Akishima, Tokyo, Japan). Gels were formed as described above and spotted onto glow-discharged carbon-coated copper Formvar grids (Ted Pella, Redding, CA). Excess liquid and gel were removed with paper or parafilm (Whatman, Maidstone, United Kingdom). Grids were submerged in 1% uranyl acetate, then blotted and air dried for 15 min before imaging. Images of gels are representative of at least four images of each gel type.
Phenyl-sepharose chromatography
Fluorescent reporters were dissolved in 200 mM NaCl, 20 mM HEPES [pH 7] and loaded into high-prep phenyl FF 1 mL-capacity phenyl-sepharose columns (GE Healthcare Lifesciences, Pittsburgh, PA) equilibrated with three volumes of 200 mM NaCl, 20 mM HEPES [pH 7]. For elution, flow rates were set to 1 mL/min with 0.5 mL fraction volumes. Fraction concentrations are representative traces of elution profiles.
Results
To determine the relevance of charged amino acids in FG domains, we chose the yeast nucleoporin Nsp1 as a model system because it is essential in Saccharomyces cerevisiae (32) and contains a repeating subsequence (284–553) with a high density of charge (Fig. 1 A). Sequence analysis of 15 repeats of FG domains in Nsp1284–553 revealed a nonuniform distribution of charge in the sequence space separating FG sequences (Fig. 1 B). Cationic residues appear near the center and edges of the repeat, whereas anionic amino acids reside at least three positions away from FSFG sequences. Moreover, several highly conserved lysine (K) residues are situated 2–3 amino acids away from the FG sequence, at positions 6 and 17, and a conserved glutamic acid (E) appears at position 9 (Fig. 1 A). The nonuniformity in conserved charge distributions and amino acid identity suggests that the charge and biochemical properties of amino acid side chains may play a role in governing how neighboring FG sequences respond to environmental substrates.
Figure 1.
Identification of conserved repeat sequences and design of simplified FG self-assembling peptides. (A) Shown here is a conserved sequence identification of the C-terminal end of the essential yeast nucleoporin Nsp1. The letters in the top position indicate the most conserved residue at each location and represent the consensus sequence. (B) Shown here is conservation of charge between FG sequences in Nsp1 repeats. Positive or negative values refer to the presence of cationic or anionic residues, respectively. Amino acid numbers correspond to position in the consensus repeats, as in Fig. 1A. (C) Depicted here are designed peptides consisting of 14 amino acids with the sequence structure FSFGAXAAXAFSFG, where X represents the substituted amino acids K, E, or S. These peptides were used to determine how the presence of charge affects FG-mediated selectivity. (D) Depicted here are designed peptides in which K is moved immediately adjacent to (FGK) or placed three amino acids away (FGA2K) from FSFG domains to test how the spatial localization of charge influences FG selectivity. (E) Depicted here is a class of peptides in which K is substituted with R and E is substituted with D to determine how biochemistry affects FG-mediated selectivity and self-assembly.
To test how the type and placement of charge as well as side-chain chemistry of neighboring amino acids affect FG function, we synthesized the consensus Nsp1 peptide sequence and rationally designed 14-amino acid variations of it. All engineered peptides consisted of two terminal FSFG sequences with neighboring charged or neutral amino acids. To establish how the presence of charge affects FG-mediated selectivity, we designed the peptide sequence FSFGAXAAXAFSFG, where X is K, E, or a neutral serine (S) (Fig. 1 C). Because K is the most conserved residue in this wild-type Nsp1 subsequence, we use the K-containing peptide (the FGAK peptide) as the reference for all other experimental comparisons. To determine whether the substrate-binding properties of FG domains depend upon the type of neighboring charge, we synthesized the anionic peptide FGAE, and as a control the neutral FGAS peptide in which each K was converted to S. To test how the positioning of K relative to the FG sequence affects FG function, we designed two variants of FGAK in which the K was placed directly adjacent to the FG domain or separated by two alanine (A) residues (peptides FGK and FGA2K, respectively; Fig. 1 D). Last, because only certain charged amino acids such as K and E exhibit 100% conservation at positions 6, 9, and 17 (Fig. 1 A), we hypothesized that amino acid biochemistries may also regulate FG-based molecular recognition. To test how the chemical structures of amino acid side chains affect the structure and function of FG domains, we designed a third class of peptides in which K was replaced with cationic arginine (R; FGAR) and E was replaced with anionic aspartic acid (D; FGAD) (Fig. 1 E). With these 14-amino acid nucleoporin-based peptides, we evaluated how single amino acid substitutions alter the selective binding and self-assembling capabilities of individual FG domains.
Characterizing the effect of charge on FG-mediated self-assembly
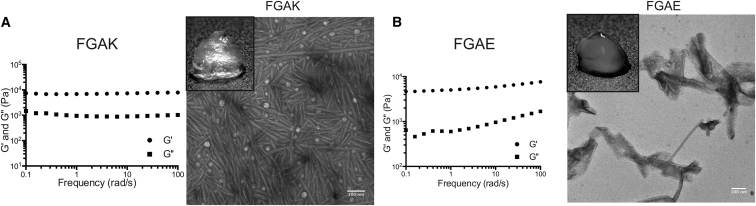
Before studying selective recognition by FG-based peptides, we first tested whether they form gels. We used a concentration of 2% (w/v) for each of the peptides, which corresponds to 28 mM FG sequence, a value that is well within the range estimated for densely packed NPCs (18, 33, 34). To quantify gelation, we measured the stiffness of the resulting material using small amplitude oscillatory frequency sweeps. We report the storage (G′) and loss (G″) moduli of the peptide solution (Fig. 2). A gel forms when G′ > G″, which indicates successful self-assembly of a stable network of peptides. The consensus peptide sequence of Nsp1 (Fig. 1 A) was unable to form a gel and flowed when inverted (data not shown), so it was not suitable for further analysis.
Figure 2.
FGAK and FGAE self-assembly. Frequency sweeps of (A) FGAK gel and (B) FGAE gel, with G′ (storage) and G″ (loss) moduli reported at 2% (w/v). (Insets) Shown is transmission electron microscopy of self-assembled peptides and macroscopic gel. For both FGAK and FGAE peptides, G′ > G″, indicating gel formation. The replacement of K with E alters the self-assembly properties of FG sequences to form different structures and gels with varying stiffness.
For the engineered FG sequences, we established that the reference peptide FGAK forms a hydrogel with a stiffness of 104 Pa across the frequencies tested (Fig. 2 A). To ensure that the FG sequences were responsible for the self-assembly process, rather than the high density of A residues, which can promote stable self-assembly (35), we converted F to S (SGAK). With this substitution, the peptide remained in the aqueous phase and no longer exhibited a dominant storage modulus (Fig. S4 A), suggesting that the F within the designed peptides provides the necessary hydrophobic interactions for gelation, as for intact FG nucleoporins (9, 10). Reversing the charge via an E (FGAE peptides) revealed that the gel-forming properties are maintained with stiffness 5–9 KPa at the frequencies tested (Fig. 2 B). To determine whether the charge is responsible for maintaining the hydration of the gel, we compared the material to the solution of neutral FGAS peptides. Without the charge, FGAS precipitated out of solution (Fig. S4 B), indicating that the presence of charge is essential in maintaining a hydrated network of FG sequences. However, it appeared that too much charge could prevent gelation, presumably by increasing the solubility, as with the consensus sequence peptide. Without electrostatic repulsion in the gels, the network collapses and forms a precipitate. Taken together, our results suggest that the sequence adjacent to the FG sequence encodes information that impacts self-assembly and substrate binding by FG domains.
Binding of NTF2 is asymmetric in cationic and anionic FG-based gels
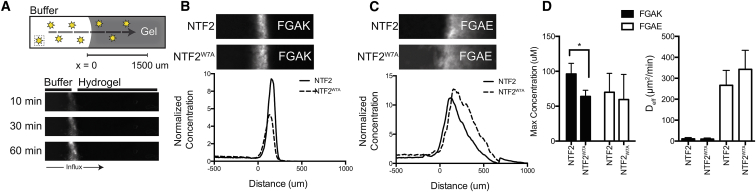
To determine whether FG-based peptide gels reconstitute the selective binding properties of native FG nucleoporins, we selected NTF2 as a model receptor because it contains an essential tryptophan (W7) that is required for binding FG domains. We replaced W with an A (NTF2W7A) to ablate FG-binding capabilities (14), enabling us to test whether the FG domains are available in the gel for NTR binding. As cationic native and engineered nucleoporin gels are predicted to bind and facilitate the selective transport of native NTRs such as NTF2 (21, 22), we first determined whether the positively charged FGAK gel preferentially selected for NTF2 compared to the NTF2W7A mutant. To prepare gels for selective transport assays, we dissolved FGAK peptides at 2% (w/v) as before and loaded the material into capillaries. Fluorescently labeled NTF2 or NTF2W7A was then injected into the capillary and sealed to create a 1D diffusion chamber; the diffusion profile was monitored for up to 5 h at 1-min intervals (Fig. 3 A).
Figure 3.
Molecular selectivity of the native transport receptor NTF2 in FGAK and FGAE gels. (A) Top, a schematic of a slab 1D transport system within a sealed capillary, which provides no flux boundary conditions during the diffusion of fluorescent molecules (stars). Bottom, the interface of the gel is reflected by the increase in fluorescence signal between the buffer and hydrogel. (B and C) Depicted here is transport of representative fluorescently labeled NTF2 and NTF2W7A proteins into FGAK and FGAE gels at 5 h. FGAK is able to preferentially recognize NTF2 over NTF2W7A mutant whereas FGAE gels are unable to. (D) Left, shown here is maximum concentration of labeled proteins at the interface. Right, shown here is effective diffusion coefficients of labeled proteins. Error bars are SDs of at least three independent replicates. ∗p < 0.05, unpaired Student’s t-test. To see this figure in color, go online.
To quantify the extent to which the gels differentiated between NTF2 and NTF2W7A, we determined the maximum accumulation of the transport receptors at the gel interface and their effective diffusion coefficients inside the gel using time-evolving fluorescence profiles. Lower effective diffusivities and higher accumulation indicate stronger interactions with the gel. For a detailed description of diffusion coefficient analysis for these time series, refer to the Materials and Methods and Fig. S3. NTF2 accumulated at higher concentrations (mean ± SD: 96.2 ± 15.2 μM) at the gel interface than did the NTF2W7A mutant (63.9 ± 8.8 μM) (Fig. 3, B and D). Thus, the FGAK peptide has a binding preference for NTF2 over the NTF2W7A mutant, and appears capable of recognizing a single hydrophobic amino acid difference. Although NTF2 displayed increased enrichment at the FGAK gel interface (Fig. 3 D), its effective diffusivity did not significantly differ from that of the NTF2W7A mutant (NTF2, 10.6 ± 6.5 μm2/min; NTF2W7A 9.9 ± 4.9 μm2/min). This experiment illustrates that the minimal in vitro system does not fully reconstitute the selective transport observed in the NPC in vivo, but it enables the characterization of the first step of selective transport in native NPCs, namely, the requirements for the initial selective binding to FG-based gels, which is the focus of this work.
To test whether NTF2 binding is sensitive to charged residues neighboring the FG domain, we determined whether NTF2’s interaction with FGAE recapitulated the selective properties of FGAK, despite the reversal of charge. Fig. 3, C and D, shows that FGAE selects for NTF2 (69.9 ± 27.1 μM) and NTF2W7A (59.5 ± 36.0 μM) equally at the interface, with no significant difference in diffusion coefficients (NTF2 266.5 ± 71.1 μm2/min and NTF2W7A 342.7 ± 91.0 μm2/min). These results indicate that the FGAE gel is less effective than the FGAK gel at differentiating between the native and mutant forms of NTF2. The FGAK and FGAE gels exhibit structural differences at the microscopic level (sheets versus fibers; Fig. 2, A and B) and the macroscopic level (insets to Fig. 2, A and B). Therefore, it is possible that due to structural differences, the FG sequences are not exposed and are unavailable for binding by NTF2 in the FGAE gel. To test this hypothesis, we determined that Nile Red dye, which fluoresces within hydrophobic environments, was detected in both FGAK and FGAE gels (Fig. S5): the uncharged hydrophobic dye interacted with the FG domains within the peptide-based gels despite the structural variation, showing that hydrophobic domains are available for binding by small molecules such as Nile Red. These results suggest that K and E may regulate the selective properties of the FG sequences from a charge interaction perspective for specific classes of molecules, as opposed to simply altering the microstructure of the gel.
Presence of charge regulates selective recognition by FG domains
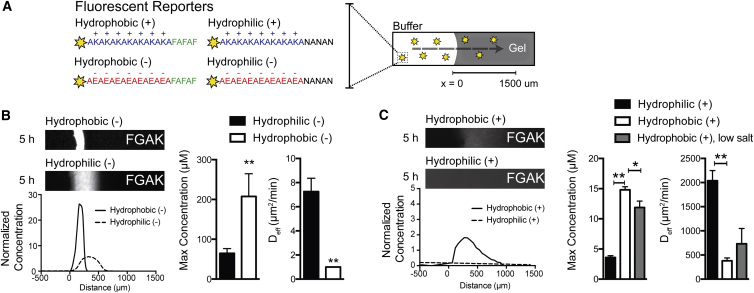
Although NTRs such as importin β (Impβ) and NTF2 have well-characterized specificity for particular FG-nup sequences, intact receptors contain multiple binding pockets with varying affinities and charge distribution, or require dimerization for function. These factors complicate the systematic analysis of how charge contributes to FG function. Hence, as a replacement for complex NTRs in our minimal in vitro model, we designed fluorescent peptide reporters with defined spatial arrangements of charged and nonpolar amino acids to systematically test the contribution of charge in hydrophobic selectivity (Fig. 4 A). The first two reporter peptides harbored three F residues, creating a hydrophobic tail for binding to FG sequences, but one contained an adjacent anionic sequence composed of E residues (termed hydrophobic (−)), whereas the other contained cationic K residues (hydrophobic (+)). As controls, we engineered two more reporters in which F residues were converted to hydrophilic asparagine (N) residues, termed hydrophilic (− or +); these control peptides should not interact with FG domains. To confirm that the synthetic hydrophobic reporters interacted with the aromatic phenyl group in FG domains in the gels independently of their charged domains, we used phenyl-sepharose columns to assay hydrophobic interactions. This assay was previously employed to isolate intact NTRs from cell lysates (12). Both hydrophobic reporters displayed longer retention times than their hydrophilic counterparts (Fig. S6), indicating that the hydrophobic reporters interact with the phenyl groups on the sepharose independent of the displayed charge.
Figure 4.
Charge affects hydrophobic molecular recognition in cationic FGAK gels. (A) Shown here is a schematic of the four fluorescent reporters with a charged domain and a hydrophilic (N) or hydrophobic (F) tail. (B) Given here are representative fluorescence images of hydrophobic (−) and hydrophilic (−) reporters in the FGAK gel, showing the increased selectivity for hydrophobic (−) reporters over hydrophilic (−) reporters. (C) Depicted here are representative fluorescence images of hydrophobic (+) and hydrophilic (+) reporters, which contain a cationic tail, into FGAK gel showing the increased selectivity for hydrophobic (+) reporters over hydrophilic (−) reporters. A low-salt condition (20 mM NaCl) was added for the hydrophobic (+) reporters to show how electrostatic screening modulates hydrophobic interactions for hydrophobic (+) reporters. For (B and C), error bars are SDs of at least three independent replicates. ∗p < 0.05 and ∗∗p < 0.01, unpaired Student’s t-test. To see this figure in color, go online.
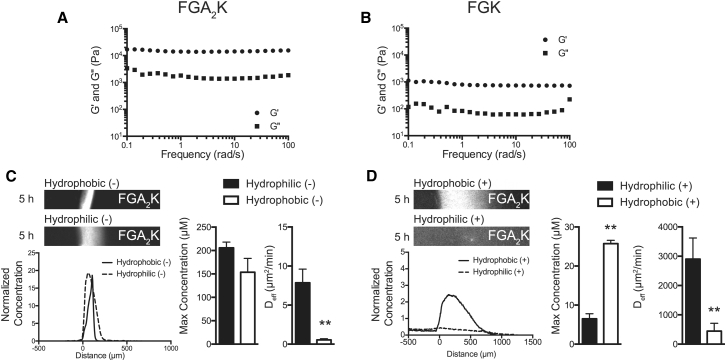
We used the FGAK gels to investigate whether the two hydrophobic fluorescent reporters underwent differential uptake into the gel. The cationic FGAK gel interacted with the hydrophobic (−) reporters, and accumulated inside the gel. The maximum concentration of peptide at the interface was approximately threefold higher for the hydrophobic (−) reporter (64.6 ± 12.1 μM) than the hydrophilic (−) reporter (207.6 ± 57.2 μM) (Fig. 4 B), which does not contain a hydrophobic tail, suggesting that FG sequences in FGAK recognize reporter F residues when the reporter is negatively charged. The contribution of hydrophobic interactions is further corroborated by the strong reduction in the effective diffusion coefficient from 7 μm2/min for hydrophilic (−) to <1 μm2/min for hydrophobic (−) (Fig. 4 B), indicating that the hydrophobic interactions induce tighter binding in the context of electrostatic attraction. We note that the interactions are so strong that the hydrophobic (−) reporters essentially do not diffuse over the 5-h period analyzed, suggesting that the binding is irreversible. In contrast, when the reporters contained a cationic tail (Fig. 4 C), the hydrophobic (+) and hydrophilic (+) reporters diffused into the gel with coefficients of 381.0 ± 57 μm2/min and 2038.0 ± 211.8 μm2/min, respectively. The hydrophobic (+) reporters bound 1.5 times above the original bath concentration at the interface (14.81 ± 0.5 μM), whereas the hydrophilic (+) reporters equilibrated (3.6 ± 0.3 μM) with no discernible partitioning (Fig. 4 C). Thus, hydrophobic interactions can occur in electrostatically repelling environments, but the interactions are much weaker than those detected with the hydrophobic (−) reporters. Moreover, lowering the salt concentrations from 200 mM NaCl to 20 mM (to decrease electrostatic screening and to increase electrostatic repulsion) decreased the accumulation of hydrophilic (+) reporter at the interface (11.9 ± 1.1 μM), whereas the diffusion coefficient (732 ± 317.7 μm2/min) trended upward (Fig. 4 C), indicating that repulsion weakens the overall strength of the hydrophobic interactions with the FG domains. These results show that the K residues in FGAK gels distinguish between two substrates that contain the same hydrophobic domain but with different surrounding charge types. In particular, K allows for increased binding to hydrophobic domains with neighboring anionic residues.
Spatial localization of lysine affects FG-mediated self-assembly and binding selectivity
Our analysis of the Nsp1 repeat consensus sequence (Fig. 1, A and B) suggests that the conserved location of K within 2–3 amino acids of the FG sequence may be relevant for FG-mediated molecular recognition. To characterize this relationship, we tested whether placing a K immediately adjacent to the FG sequence (FGK) or moving it three amino acids away (FGA2K) affected the sequence’s selectivity compared to the original arrangement in the FGAK peptide. Whereas FGA2K peptides readily formed a stiff material and could be tested for selective uptake, the FGK peptide gel was two orders-of-magnitude less stiff (Fig. 5, A and B), flowed when inverted, and dispersed when fluorescent reporters were loaded (data not shown). We therefore focused our comparison on diffusion and accumulation in FGAK and FGA2K gels. The FGAK gel selectively enriched for the hydrophobic (−) reporter but failed to uptake the hydrophobic (+) reporter to the same degree (Fig. 4, B and C). The effective diffusivity of the hydrophobic (−) (0.5 ± 0.14 μm2/min) reporter in the FGA2K gel was an order-of-magnitude lower than that of the hydrophilic (−) reporter (7.9 ± 1.7 μm2/min) (Fig. 5 C), showing that the combination of hydrophobic and electrostatic attraction synergizes for strong binding. However, the maximum accumulation at the interface of the two anionic reporters was similar (153. 6 ± 29.6 μM for hydrophobic (−) versus 205.7 ± 12.2 μM for hydrophilic (−)) (Fig. 5 C), indicating that the FGA2K gel is unable to differentiate between the two reporters to the same degree as the original FGAK gel. For cationic reporters, the hydrophobic (+) peptide accumulated within the gel (25.8 ± 0.8 μM) and displayed an order-of-magnitude lower effective diffusivity (444.3 ± 267.7 μm2/min) than the hydrophilic (+) reporter (6.5 ± 1.3 μM and 2902 ± 719.8 μm2/min, respectively) (Fig. 5 D). In addition, more hydrophobic (+) reporter accumulates in FGA2K gels than in FGAK gels (Fig. 5 D). These data show that placing K within two amino acids of an FG domain, approximately corresponding to the 1 nm Debye length at physiologically relevant salt concentrations, enables hydrophobic selection of substrates with opposite charge, whereas placing K three amino acids away reduces the contribution of electrostatic interactions to hydrophobic FG-mediated selectivity. As a result, by increasing the distance between charged residues and FG sequences, the FG sequences become less dependent on the electrostatic profile surrounding the hydrophobic substrate during selective transport.
Figure 5.
Effects of spatial localization of charge on FG-mediated self-assembly and selectivity. Frequency sweeps of (A) FGA2K and (B) FGK gels with G′ (storage) and G″ (loss) moduli are reported at 2% (w/v). (C) Shown here are representative fluorescence images of hydrophobic (−) and hydrophilic (−) reporters in a FGA2K gel, showing the similarities between the selectivity of the two reporters. (D) Depicted here are representative fluorescence images of hydrophobic (+) and hydrophilic (+) reporters, which contain a cationic tail, in a FGA2K gel showing the ability of FGA2K gels to select for hydrophobic (+) reporters independent of electrostatic repulsion. Error bars are SDs of at least three independent replicates. ∗∗p < 0.01, unpaired Student’s t-test.
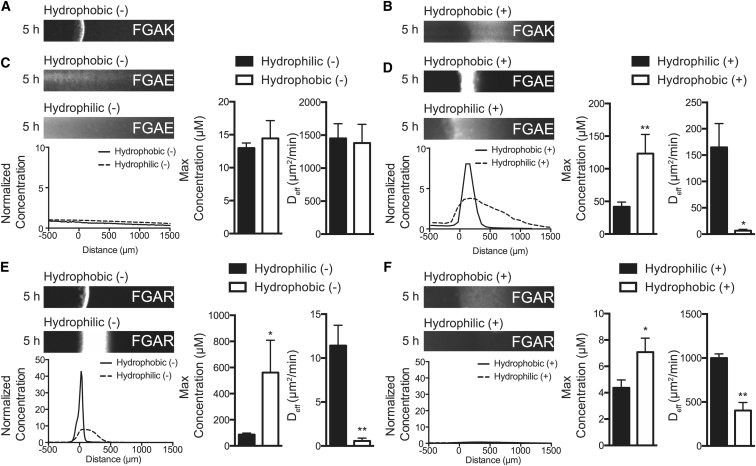
Glutamic acid reverses the selectivity of FG domains
Because the K residues in the FGAK gel help FG sequences differentiate between reporters containing anionic or cationic hydrophobic domains, we next asked whether FG-mediated recognition could be reversed by substituting anionic E residues. The selectivity of FGAE gels for the same class of fluorescent reporters was indeed reversed from that of FGAK gels (Fig. 6, A–D). Selectivity was a function of both hydrophobic and electrostatic interactions, as the hydrophilic (+) reporters accumulated threefold less at the interface (41.8 ± 7.1 μM) than did the hydrophobic (+) reporters (123.2 ± 29.32 μM) (Fig. 6 D). Moreover, the diffusion coefficient of the hydrophobic (+) reporter (6.6 ± 2.6 μm2/min) was an order-of-magnitude lower than that of the hydrophilic (+) reporter (160.8 ± 32.7 μm2/min) (Fig. 6 D). Conversely, neither the hydrophobic (−) nor the hydrophilic (−) reporter interacted significantly with the gel; their similar effective diffusion coefficients (1381.0 ± 282.0 and 1451.0 ± 223.0 μm2/min, respectively) (Fig. 6 C) show that electrostatic repulsion minimizes hydrophobic interactions with FG sequences.
Figure 6.
Substituting K with E reverses selectivity of FGAK gels, whereas replacement of K with R maintains selectivity similar to that of FGAK gels. Shown here are representative (A) hydrophobic (−) and (B) hydrophobic (+) fluorescence images in FGAK gels. (C) Shown here are representative fluorescence images of hydrophobic (−) and hydrophilic (−) reporters in an FGAE gel with the corresponding concentration profiles showing the reversal of selective recognition when compared to FGAK gels with the same reporters. In FGAE gels, anionic reporters do not interact at the gel interface. (D) Shown here are representative fluorescence images of hydrophobic (+) and hydrophilic (+) reporters in an FGAE gel, showing the reversal of selective recognition when compared to FGAK gels. In FGAE gels, the hydrophobic (+) reporter binds more significantly at the interface compared to the hydrophilic (+) reporter. (E) Depicted here are representative fluorescence images of hydrophobic (−) and hydrophilic (−) reporters in an FGAR gel, showing the similarity of selective recognition to that of FGAK gels. (F) Given here are representative fluorescence images of hydrophobic (+) and hydrophilic (+) reporters in an FGAR gel, showing the similarity of selective recognition to that of FGAK gels. Error bars are SDs of at least three independent replicates. ∗p < 0.05 and ∗∗p < 0.01, unpaired Student’s t-test.
One general concern in synthesizing anionic peptides is residual TFA protonating anionic groups such as E. To ensure that the reporters were not neutralized in our buffering conditions, we synthesized neutral reporter peptides hydrophilic (n) and hydrophobic (n) (Fig. S7). In diffusion experiments with FGAK and FGAE gels, the neutrally charged reporters aggregated in solution and interacted minimally with the gels (Fig. S7), indicating that the anionic peptides are not neutralized by the residual TFA and indeed carry a net negative charge. Taken together, the data in Fig. 6 and Fig. S7 suggest that charge proximal to FG sequences can tune hydrophobic selectivity, and that charge is essential in determining the hydrophobic moieties recognized by FG sequences.
Amino acid side-chain chemistry is an additional regulator of FG function
Last, we tested how the side-chain chemistry of charged amino acids affects the self-assembly and molecular recognition of FG sequences. In Nsp1, the predominant cation is K rather than R (Fig. 1 A), suggesting that the two cations may not function equally. Similarly, E is highly conserved at position 9, whereas D is not as well conserved throughout the repeats (Fig. 1 A). To test the importance of side-chain chemistry, we determined in the diffusion assay that FGAR gels have uptake properties similar to those of FGAK gels (Fig. 6, A and E). The hydrophobic (−) reporter accumulated (561.1 ± 247.7 μM) and interacted with the interface of the FGAR gels (0.6 ± 0.3 μm2/min), whereas the hydrophobic (+) reporter did not (7.1 ± 1.1 μM and 403.7 ± 90.2 μm2/min, respectively) (Fig. 6, E and F). These data suggest that from an electrostatic standpoint, R is just as capable as K in helping an FG sequence differentiate between hydrophobic substrates. However, mechanically, FGAR forms gels with an approximate stiffness of 2000 Pa throughout the frequency sweep (Fig. S8 A), fourfold more compliant than FGAK gels (Fig. 2 A). The differences in the mechanical properties of the FGAK and FGAR gels suggest that the K and R residues may predominantly affect the structural self-assembly of FG domains. Transmission electron microscopy revealed that FGAR peptides (Fig. S8 B) form different structures than FGAK peptides (Fig. 2 A).
FGAD did not form a gel at the standard 2% (w/v), formed no repeating structures (Fig. S8, C and D), and inverted when flowed (data not shown), suggesting that the small chemical differences between E and D transition the material from a selective gel to a viscous solution. Taken together, these data show that in addition to net charge, amino acid side-chain properties such as their inherent hydrophobicity, side-chain sterics, and charge distribution all may be important parameters that determine hydrophobic FG-mediated interactions.
Tryptophan interactions are similarly modulated by electrostatics
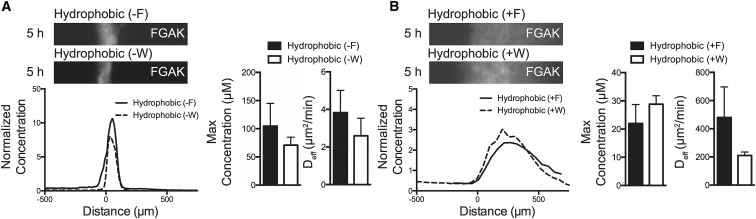
An essential question in the selectivity of FG domains is how generalizable the tunability of hydrophobic selectivity is to natural aromatic amino acids other than F. In native NTRs such as NTF2 and other transport receptors, tryptophan (W) is commonly used for the transient hydrophobic interactions required for transport. To test whether an electrostatic dependence persists when F is converted to W, we constructed the reporters hydrophobic (+W) and hydrophobic (−W) from the original hydrophobic F reporters (termed +F and −F, respectively, for the remainder of the article). The W-containing reporters were modulated by electrostatic interactions in a similar manner to F-containing reporters. The maximum accumulated fluorescence at the interface and diffusion did not significantly differ between the hydrophobic −F and −W reporters (Fig. 7 A). The cationic reporters exhibited similar maximum interface properties, but the +W reporter showed a slight and nonsignificant trend toward slower diffusion than the +F reporter (+F: 479.9 ± 217.2 μm2/min; +W: 210.5 ± 24.6 μm2/min) (Fig. 7 B), indicating that although both aromatic residues can be modulated by electrostatic interactions, the identity of the hydrophobic residue has a small effect on transport in the gel. These results reveal that in the NPC, the phenomenon of electrostatic interactions regulating hydrophobic interactions may be generalizable to aromatic residues other than F; moreover, selective transport may also be tuned by the identity of the nonpolar amino acid.
Figure 7.
Electrostatic interactions modify hydrophobic recognition of W-containing reporters similar to F-containing reporters in FGAK gels. Comparisons between (A) hydrophobic (−F) and hydrophobic (−W) reporters and (B) hydrophobic (+F) and hydrophobic (+W) reporters in FGAK gels show that replacing F with W does not alter hydrophobic molecular recognition for FGAK gels. Representative fluorescence images and corresponding concentration profiles are shown. Error bars are SDs of at least three independent replicates. No significant differences were detected via Student’s t-test.
Discussion
This investigation demonstrates how the environment surrounding FG sequences tunes the hydrophobic interactions necessary for correct molecular recognition of hydrophobic substrates. Here, we have explicitly shown how the presence, placement, and type of charged amino acid are all essential parameters for FG function in gels. Translating concepts that are well established in the peptide field enabled us to establish how even simplified variants of the Nsp1 consensus sequence can encode complex molecular recognition. The data reported here support previous theoretical predictions that electrostatic interactions may be an important regulator of NPC selectivity (22) and sequence observations that the bias in the net charge of NTRs may be important in transport through the NPC (21). Moreover, recent reports on how charge influences interactions with hydrophobic surfaces (8, 36) emphasize that the interplay between electrostatics and hydrophobicity may be fundamental for various biological functions such as molecular recognition and DNA packing (37, 38).
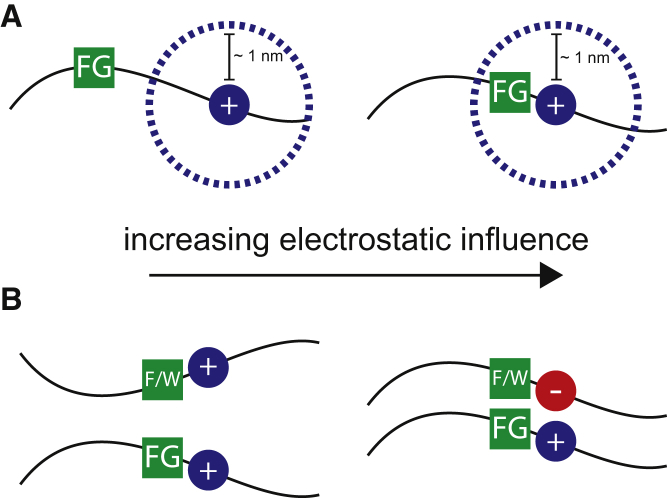
For the nuclear pore field, this investigation shifts the spotlight from how hydrophobic effects determine NPC selectivity to how electrostatics contribute to this selectivity (9, 11, 12, 39). The results described here provide insight into how individual hydrophobic FG sequences may distinguish a particular subset of hydrophobic substrates using a combination of electrostatic and hydrophobic domains that are in close proximity to each other (Fig. 8). The closer the charged residues are to FG sequences, the greater the influence of charge on FG self-assembly (Fig. 8 A) and recognition of diffusing substrates (Fig. 8 B). As the charged residue is situated greater than the theoretical Debye length at physiological conditions, the hydrophobic and electrostatic motifs are able to act independently for molecular recognition.
Figure 8.
Schematic of the influence of electrostatics on hydrophobic interactions mediated by FG domains. (A) At physiologically relevant salt concentrations, electrostatic interactions typically have a range of ∼1 nm (Debye length, dotted lines), which determines how much a charged residue influences the FG sequence’s recognition of and binding to hydrophobic substrates. When charged residues are moved further away from the FG sequence, electrostatic interactions become less significant for FG-mediated self-assembly and selective transport. (B) Complementary charge and hydrophobic interactions enable the strongest interactions between a substrate and FG domains. Modulating charge types and the presence of hydrophobic domains can tune interactions from strong binding to free diffusion within the gel and in in vivo systems. To see this figure in color, go online.
We recognize that although the study of engineered peptides can answer questions on how hydrophobic interactions are affected by the precise placement of charged amino acids, short-peptide gels do not recapitulate all of the unique facilitated diffusion properties of intact FG nucleoporins. This disconnect likely arises from structural differences, as short peptides form fibers and rods (Fig. 2 and Fig. S8) that are not the dominant structures observed or predicted in the native nuclear pore (7, 40). Moreover, the interactions between diffusing reporter peptides and the self-assembled gel may cause structural changes in the gel and also alter the diffusivity of the reporter, which has also been reported for Impβ in native NPCs (41). Nevertheless, this investigation of peptides provides strong evidence (Figs. 4, 5, and 6) that charge is an essential regulator of transport through the NPC by tuning the essential first step of recognizing specific hydrophobic substrates. Moreover, engineered FG-containing peptides do capture the interplay between hydrophobic and electrostatic interactions at single amino acid resolution (Fig. 5), which is not possible with currently available in vivo techniques.
Our results build on previous studies of intact nucleoporins containing multiple repeats of FG domains where charge primarily plays a structural role in the cohesion of the self-assembled matrix (40). Our results also support recent work in which Impβ was recognized to have varying binding capacities with synthetic FG-containing polyacrylamide gels depending on the charged state of the material (42). We believe that our strategy, which was inspired and informed by peptide engineering, complements other well-established methods used to understand the selective transport mechanism of NPCs, such as the in vivo minimal NPC (18, 19), gel and selectivity analysis of individual nucleoporins (9, 10, 11, 43), and binding interactions with surface-grafted nucleoporin films (40, 44, 45, 46). We expect that this rational approach from the peptide field, which focuses on conserved repeating sequences, can be extended to other biological gel systems such as mucus, byssal threads, and cartilage, where complex disordered proteins based on repeat units and reversible cross linking constitute a significant proportion of the material and dictate its function (47, 48, 49).
Author Contributions
W.G.C. and K.R. designed the research. W.G.C., J.W., and S.C.G performed the experiments. All authors contributed to writing and editing of the article.
Acknowledgments
We thank Prof. Alan Grodzinsky at the Massachusetts Institute of Technology for his valuable feedback and helpful discussions, and Felice Frankel for the images of the gels in the insets of Figure 2, A and B.
This work was supported by the Materials Research Science and Engineering Center Program of the National Science Foundation under award DMR-0819762, Defense Threat Reduction Agency under awards HDTRA1-13-1-0038 and NIH RO1 R01-EB017755, and National Science Foundation CAREER award PHY-1454673. W.G.C. was supported in part by the National Institute of General Medical Sciences/National Institutes of Health Interdepartmental Biotechnology Training Program under T32 GM008334. J.W. was supported in part by the National Science Foundation Graduate Research Fellowship Program under grant No. 1122374.
Editor: Andrew Spakowitz.
Footnotes
Wesley G. Chen and Jacob Witten contributed equally to this work.
Supporting Materials and Methods and eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31023-8.
Supporting Material
References
- 1.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 2.Stewart M., Baker R.P., Matsuura Y. Molecular mechanism of translocation through nuclear pore complexes during nuclear protein import. FEBS Lett. 2001;498:145–149. doi: 10.1016/s0014-5793(01)02489-9. [DOI] [PubMed] [Google Scholar]
- 3.Görlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 4.Ando D., Zandi R., Gopinathan A. Nuclear pore complex protein sequences determine overall copolymer brush structure and function. Biophys. J. 2014;106:1997–2007. doi: 10.1016/j.bpj.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribbeck K., Görlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghavami A., Veenhoff L.M., Onck P.R. Probing the disordered domain of the nuclear pore complex through coarse-grained molecular dynamics simulations. Biophys. J. 2014;107:1393–1402. doi: 10.1016/j.bpj.2014.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamini R., Han W., Schulten K. Assembly of Nsp1 nucleoporins provides insight into nuclear pore complex gating. PLOS Comput. Biol. 2014;10:e1003488. doi: 10.1371/journal.pcbi.1003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma C.D., Wang C., Abbott N.L. Modulation of hydrophobic interactions by proximally immobilized ions. Nature. 2015;517:347–350. doi: 10.1038/nature14018. [DOI] [PubMed] [Google Scholar]
- 9.Frey S., Görlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 10.Frey S., Richter R.P., Görlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 11.Frey S., Görlich D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009;28:2554–2567. doi: 10.1038/emboj.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribbeck K., Görlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayliss R., Littlewood T., Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J. Biol. Chem. 2002;277:50597–50606. doi: 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 14.Bayliss R., Ribbeck K., Stewart M. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 1999;293:579–593. doi: 10.1006/jmbi.1999.3166. [DOI] [PubMed] [Google Scholar]
- 15.Strawn L.A., Shen T., Wente S.R. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- 16.Grant R.P., Neuhaus D., Stewart M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1Å resolution. J. Mol. Biol. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- 17.Bednenko J., Cingolani G., Gerace L. Importin-β contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strawn L.A., Shen T., Wente S.R. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 19.Patel S.S., Belmont B.J., Rexach M.F. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 20.Ribbeck K., Kutay U., Görlich D. The translocation of transportin-cargo complexes through nuclear pores is independent of both ran and energy. Curr. Biol. 1999;9:47–50. doi: 10.1016/s0960-9822(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 21.Colwell L.J., Brenner M.P., Ribbeck K. Charge as a selection criterion for translocation through the nuclear pore complex. PLOS Comput. Biol. 2010;6:e1000747. doi: 10.1371/journal.pcbi.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagliazucchi M., Peleg O., Szleifer I. Effect of charge, hydrophobicity, and sequence of nucleoporins on the translocation of model particles through the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 2013;110:3363–3368. doi: 10.1073/pnas.1212909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando D., Colvin M., Gopinathan A. Physical motif clustering within intrinsically disordered nucleoporin sequences reveals universal functional features. PLoS One. 2013;8:e73831. doi: 10.1371/journal.pone.0073831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada J., Phillips J.L., Rexach M.F. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 26.Hinman M.B., Jones J.A., Lewis R.V. Synthetic spider silk: a modular fiber. Trends Biotechnol. 2000;18:374–379. doi: 10.1016/s0167-7799(00)01481-5. [DOI] [PubMed] [Google Scholar]
- 27.Xu M., Lewis R.V. Structure of a protein superfiber: spider dragline silk. Proc. Natl. Acad. Sci. USA. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer D.E., Chilkoti A. Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 29.Nettles D.L., Chilkoti A., Setton L.A. Applications of elastin-like polypeptides in tissue engineering. Adv. Drug Deliv. Rev. 2010;62:1479–1485. doi: 10.1016/j.addr.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright E.R., Conticello V.P. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Adv. Drug Deliv. Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 31.Crooks G.E., Hon G., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurt E.C. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4323–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denning D.P., Patel S.S., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denning D.P., Uversky V., Fink A.L. The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J. Biol Chem. 2002;277:33447–33455. doi: 10.1074/jbc.M203499200. [DOI] [PubMed] [Google Scholar]
- 35.Rathore O., Sogah D.Y. Self-assembly of β-sheets into nanostructures by poly(alanine) segments incorporated in multiblock copolymers inspired by spider silk. J. Am. Chem. Soc. 2001;123:5231–5239. doi: 10.1021/ja004030d. [DOI] [PubMed] [Google Scholar]
- 36.Chen S., Itoh Y., Aida T. Ionic interactions. Subnanoscale hydrophobic modulation of salt bridges in aqueous media. Science. 2015;348:555–559. doi: 10.1126/science.aaa7532. [DOI] [PubMed] [Google Scholar]
- 37.Migliori A.D., Smith D.E., Arya G. Molecular interactions and residues involved in force generation in the T4 viral DNA packaging motor. J. Mol. Biol. 2014;426:4002–4017. doi: 10.1016/j.jmb.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Migliori A.D., Keller N., Smith D.E. Evidence for an electrostatic mechanism of force generation by the bacteriophage T4 DNA packaging motor. Nat. Commun. 2014;5:4173. doi: 10.1038/ncomms5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hülsmann B.B., Labokha A.A., Görlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Ader C., Frey S., Baldus M. Amyloid-like interactions within nucleoporin FG hydrogels. Proc. Natl. Acad. Sci. USA. 2010;107:6281–6285. doi: 10.1073/pnas.0910163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe A.R., Tang J.H., Liphardt J.T. Importin-β modulates the permeability of the nuclear pore complex in a Ran-dependent manner. eLife. 2015;4:e04052. doi: 10.7554/eLife.04052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman A.K., Baker L.A. Synthetic hydrogel mimics of the nuclear pore complex display selectivity dependent on FG-repeat concentration and electrostatics. Soft Matter. 2016;12:9477–9484. doi: 10.1039/c6sm01689h. [DOI] [PubMed] [Google Scholar]
- 43.Labokha A.A., Gradmann S., Görlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisele N.B., Labokha A.A., Richter R.P. Cohesiveness tunes assembly and morphology of FG nucleoporin domain meshworks—implications for nuclear pore permeability. Biophys. J. 2013;105:1860–1870. doi: 10.1016/j.bpj.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eisele N.B., Frey S., Richter R.P. Ultrathin nucleoporin phenylalanine-glycine repeat films and their interaction with nuclear transport receptors. EMBO Rep. 2010;11:366–372. doi: 10.1038/embor.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zahn R., Osmanović D., Richter R.P. A physical model describing the interaction of nuclear transport receptors with FG nucleoporin domain assemblies. eLife. 2016;5:e14119. doi: 10.7554/eLife.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holten-Andersen N., Zhao H., Waite J.H. Stiff coatings on compliant biofibers: the cuticle of Mytilus californianus byssal threads. Biochemistry. 2009;48:2752–2759. doi: 10.1021/bi900018m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waller K.A., Zhang L.X., Jay G.D. Role of lubricin and boundary lubrication in the prevention of chondrocyte apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5852–5857. doi: 10.1073/pnas.1219289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose M.C., Voynow J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.