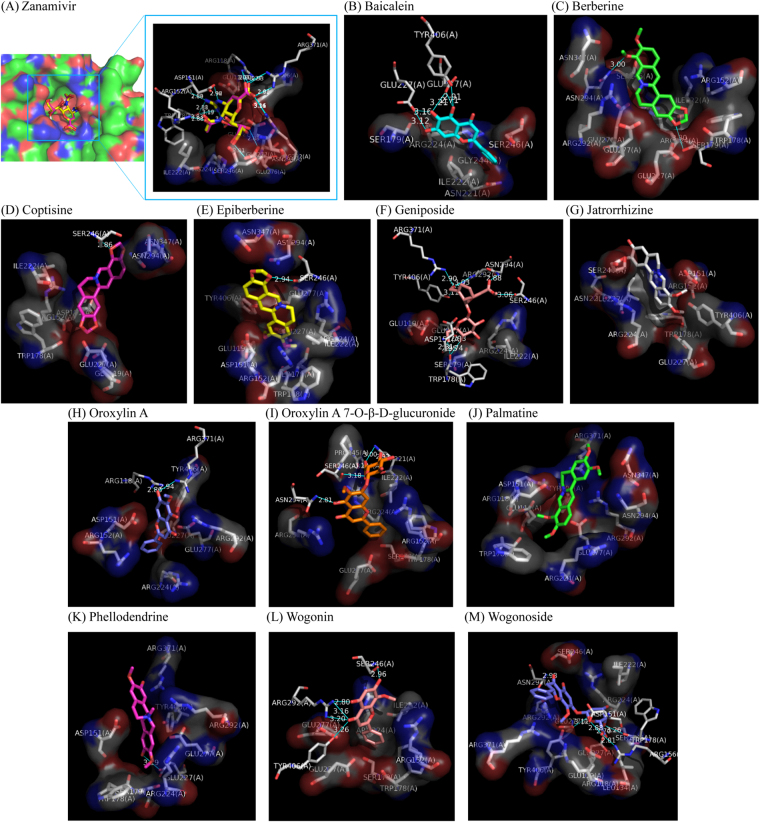
Figure 4.
Molecular docking simulation shows the favorable binding positions of potent neuraminidase-1 inhibitors from the plasma profile of Huang-Lian-Jie-Du-Tang with lower binding free energy and proper distances (more than 1 Å) to water molecules in the ligand-binding cavity of H1N1 neuraminidase (PDB ID 3B7E). The 3D diagrams display the interactions of (A) zanamivir (re-docked pose, in purple sticks; crystallographic pose, in yellow sticks), (B) baicalein (in cyan sticks), (C) berberine (in green sticks), (D) Coptisine (in purple sticks), (E) Epiberberine (in yellow sticks), (F) Geniposide (in dull-red sticks), (G) Jatrorrhizine (in white sticks), (H) Oroxylin A (in blue sticks), (I) Oroxylin A 7-O-β-D-glucuronide (in ginger sticks), (J) Palmatine (in green sticks), (K) Phellodendrine (in purple sticks), (L) Wogonin (in dull-red sticks), and (M) Wogonoside (in blue sticks) to H1N1 neuraminidase with labeled amino residues responsible for generating binding free energy. Surfaces represent amino residues responsible for hydrophobic contacts with ligands. Blue lines with respective distances represent H-bonding between ligands and amino acid residues.