Abstract.
To create tumor “habitats” from the “signatures” discovered from multimodality metabolic and physiological images, we developed a framework of a processing pipeline. The processing pipeline consists of six major steps: (1) creating superpixels as a spatial unit in a tumor volume; (2) forming a data matrix containing all multimodality image parameters at superpixels; (3) forming and clustering a covariance or correlation matrix of the image parameters to discover major image “signatures;” (4) clustering the superpixels and organizing the parameter order of the matrix according to the one found in step 3; (5) creating “habitats” in the image space from the superpixels associated with the “signatures;” and (6) pooling and clustering a matrix consisting of correlation coefficients of each pair of image parameters from all patients to discover subgroup patterns of the tumors. The pipeline was applied to a dataset of multimodality images in glioblastoma (GBM) first, which consisted of 10 image parameters. Three major image “signatures” were identified. The three major “habitats” plus their overlaps were created. To test generalizability of the processing pipeline, a second image dataset from GBM, acquired on the scanners different from the first one, was processed. Also, to demonstrate the clinical association of image-defined “signatures” and “habitats,” the patterns of recurrence of the patients were analyzed together with image parameters acquired prechemoradiation therapy. An association of the recurrence patterns with image-defined “signatures” and “habitats” was revealed. These image-defined “signatures” and “habitats” can be used to guide stereotactic tissue biopsy for genetic and mutation status analysis and to analyze for prediction of treatment outcomes, e.g., patterns of failure.
Keywords: superpixel, tumor habitat, clustering analysis
1. Introduction
Tumor heterogeneity presents a great challenge for diagnosis, staging, treatment, and therapeutic response assessment. As an example, glioblastoma (GBM), an aggressive primary brain tumor in adults, exhibits profound inter- and intratumoral heterogeneity. Genomic analysis has classified GBM tumors into molecular subtypes that have different outcomes following standard chemoradiation therapy.1,2 In addition to having a dominant genomic subtype, tumors have intratumoral heterogeneity, such as having regions with distinct genomic subtypes. Imaging is a noninvasive tool to assess tumor heterogeneity. Various imaging techniques, from conventional magnetic resonance imaging (MRI), such as contrast-enhanced T1-weighted and T2-weighted fluid attenuation inversion recovery (FLAIR) images, to advanced metabolic and physiological imaging, e.g., proton MR spectroscopy, cerebral blood volume (CBV), quantitative vascular leakage measurement, conventional and high -value diffusion-weighted MRI (DW-MRI) and 11C-methionine (MET) positron emission tomography (PET), have all shown value for prognosis and prediction of failure following chemoradiation.3–13 However, abnormalities captured by these imaging modalities vary spatially from region-to-region in a single tumor. For example, the tumor subvolumes with elevated CBV (that is an established imaging biomarker for prognosis) and with hypercellularity (detected by high -value diffusion images and predicting progression and survival) are largely distinct.14 Other multiparametric images demonstrate spatial similarity and parameter correlation in patients with GBM.
Currently, methods and tools are lacking to efficiently integrate and analyze multimodality images to create tumor “signatures” and then link them to tumor “habitats” at specific locations. Tumor signatures have been discovered to characterize properties of the entire tumor in genomic and radiomic studies. For instance, in the genomic analysis, a hierarchical clustering has been used to identify patterns from thousands of gene expressions data of a tumor.15–18 In the radiomics analysis that often creates hundreds of features, the clustering analysis is used to identify imaging phenotype patterns and then to associate them with prognostic data.19–21 However, considering that imaging by nature provides spatial information, the image-defined signatures of a tumor, after discovery, should be decoded back to the spatial domain to create specific signature-defined regions as habitats. These image signature-defined tumor habitats can be used to guide stereotactic biopsy to determine their associations with types and subtypes of genomic and mutation status, and compare with locations of treatment failure. Ultimately, the validated image-defined tumor habitats with histopathology, gene expression, and mutation status could help optimize treatment strategies for individual patients.
This study aimed to develop a framework of the processing pipeline to create tumor habitats through the discovery of signatures from multimodality imaging. This analysis includes four levels. The first level of analysis is to discover the major associative signatures of the multimodality images of a tumor. The second level of analysis is to sort the superpixels (that are used as a basic spatial unit) according to the major signatures found at the previous step. The third level of analysis is to map and mark the superpixels that have the same or overlapped signatures to the image space to create the tumor habitats. The forth level of analysis is to discover the subgroup patterns of the tumors. For a proof-of-concept, we applied this framework of analysis to a set of multimodality parametric images (including 10 parameters) acquired in a group of patients treated on a prospective protocol for newly diagnosed GBM to identify the signatures and habitats. In addition, we applied the processing pipeline to the image data from another group of patients, who had GBM to test the generalizability of the pipeline and to demonstrate the clinical association of the image-defined signatures and habitats.
2. Method
2.1. Processing Pipeline
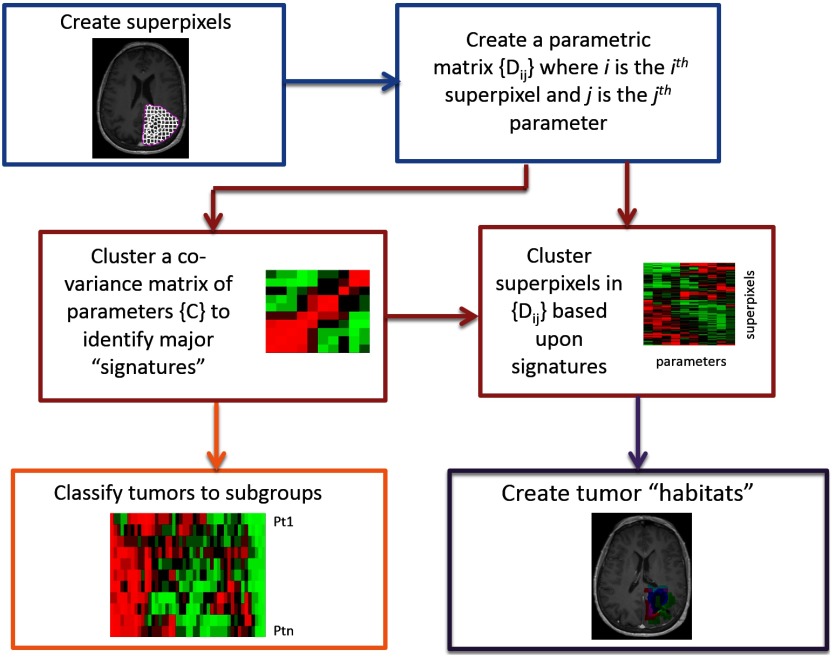
The framework of the processing pipeline to discover the image-defined tumor signatures and create the habitats is shown in Fig. 1. It includes six major steps. First, superpixels as basic spatial units are created in a tumor volume to represent the image parameters for the next image processing. Second, a data matrix per tumor is created to contain multimodality image parameters defined on superpixels. Third, a covariance or correlation coefficient (Pearson or Spearman) matrix of the image parameters is computed and analyzed by hierarchical clustering to identify major tumor signatures. Fourth, the parameter matrix is reorganized by hierarchical clustering of the superpixels and reordering the parameters according to the major clusters found in step 3. Fifth, habitats are created in the image space from the superpixels associated with the major signatures. Sixth, the covariances or correlation coefficients of each pair of image parameters are pooled together from all patients and analyzed by a hierarchical analysis to identify subgroup patterns of the tumors. Finally, these image-defined signatures and habitats can be analyzed for prediction of treatment outcomes, e.g., comparing the habitats with patterns of failure, and guidance for stereotactic tissue biopsy for genetic and mutation status analysis. In the following paragraphs, each step of the processing pipeline is described in detail.
Fig. 1.
Flowchart of the processing pipeline for discovery of tumor image signatures and habitats.
2.1.1. Superpixel creation
Superpixels are perceptually meaningful atomic segments in an image and created as a basic unit to replace the rigid structure of the pixel grid for image processing tasks.22 Superpixels are distributed regularly in the image but with desirable variations in the size and the boundary corresponding to natural variations in the image. Superpixels provide convenient primitives for image feature extraction and reduce redundancy and uncertainty in the image. The size and compactness of the superpixel can be tuned according to the uncertainty in geometric alignment of the multiple images, the image-parameter variation of interest, and the intrinsic spatial resolution of the images. Among many superpixel algorithms in the literature,22 we selected the simple linear iterative clustering (SLIC) for its superior performance, simplicity, and availability of its source code.23 SLIC adapts -means clustering to generate superpixels from intensity and coordinate information of each pixel. It provides two parameters to adjust the desired number of superpixels and their compactness, which control the size and shape of the superpixel. Superpixels can be created from one image parameter or a set (vector) of image parameters and then applied to the same or other images to extract image parameters for subsequent analysis. The extracted image parameters from each tumor can be presented by a parameter matrix , where for superpixel () and for image parameter ().
2.1.2. Discovery of the superpixel-parameter signatures
In general, there are hundreds to thousands of superpixels generated per tumor and approximately 10 or fewer image parameters. It is a challenging task to identify the parameter signatures in a superpixel-parameter matrix that pools data from all tumors. There are many possibilities for clustering and leaf ordering. Thus, intermediate steps are added in our processing pipeline. A covariance or correlation matrix of image parameters is created from the parameter matrix for each tumor. A hierarchical clustering with an optimal leaf ordering is applied to the correlation matrix of each tumor to reveal the subgroups of parameters that are correlated with each other.15,24 In this step of the analysis, the parameter similarities are analyzed at the tumor level but not at the superpixel level, which helps to identify the major patterns and the order of the parameters. For hierarchical clustering and heatmap generation, “clustergram”25 function in MATLAB (MathWorks, Natick, Massachusetts) was used. The function performs hierarchical clustering with the optimal leaf ordering24 on the data and creates dendrograms and heatmaps of the resulting clusters. At each step of the process, the two clusters separated by the shortest Euclidean distance based upon the complete-linkage criterion are combined. The process is stopped until all data are combined into a single cluster.
In the next step of the tumor signature discovery, the superpixel-parameter matrix is analyzed. First, the parameter order in the matrix is reorganized according to the major signatures or the orders that are found in the covariance or correlation matrix analysis. Then, the same hierarchical clustering with the optimal leaf ordering is applied to the superpixels in the matrix . By this method, the major parameter signatures in the superpixel-parameter matrix are retained. The resulting clusters of each tumor displayed in the heatmap, which is called the clustergram thereafter, show clusters of superpixels with similar, dissimilar, or overlapped parameters. Then, a habitat is created in the image space from a cluster of superpixels with similar parameter(s) and color-coded for visualization.
Finally, to discover subtypes of tumors, a matrix that contains all pairs of correlation coefficients of image parameters arranged by row and patients by column is created and clustered using the clustergram function in MATLAB.
2.2. Materials
For a proof-of-concept, we applied this processing pipeline to 11 patients with newly diagnosed GBM, who were imaged by conventional MRI, advanced MR sequences, and 11C-MET PET prior to chemoradiation therapy. To test the generalization of the processing pipeline and demonstrate the clinical association of image-defined signatures and habitats, we applied the process to another dataset that consisted of 10 patients with GBM, who were scanned on different scanners and had recurrence data available for comparison.
2.2.1. Patients
The first dataset consisted of 11 patients, who were enrolled on an IRB approved prospective phase II study for adult patients with newly diagnosed GBM (NCT02805179) in which patients were treated with dose-escalated chemoradiation (75 Gy simultaneous integrated boost) targeted against an identifiable area of abnormality detected on high -value DW-MRI. All patients were enrolled after maximal safe resection confirming pathology, and underwent postoperative, preradiotherapy (pre-RT), multiparametric MRI as well as 11C-MET PET. Four patients underwent gross total resection of all enhancing tumor as seen on gadolinium (Gd) enhanced T1-weighted MRI. Three of the 10 patients with available -methylguanin-DNA-methyltransferase (MGMT) status had MGMT methylation.
The second dataset consisted of 10 patients, who were enrolled on a different IRB approved prospective phase I/II radiation dose-escalation clinical trial for adult patients with newly diagnosed GBM.12,13 The trial was open in 2002, and the recruitment was completed in 2007. The retrospective data analysis was approved by the IRB. All patients underwent postoperative, pre-RT conventional MRI and MET PET. Five patients had gross total resection of all enhanced tumors. Two of the nine patients with available MGMT status had MGMT methylation. All 10 patients had tumor recurrence, determined by the multidisciplinary term according to the clinical protocol, and had available recurrence tumor contours, defined on the post-Gd T1-weighted images, to compare with image-defined signatures and habitats pre-RT.
2.2.2. Image acquisition
All patients in the first dataset had pre-RT MRI scans performed on a 3.0-T scanner (Skyra, Siemens). Conventional clinical 2-D T2-weighted FLAIR images, and 3-D pre- and post-Gd T1-weighted images were acquired. The 2-D diffusion-weighted images were acquired using a readout segmented (RESOLVE) echo planar sequence, which permits the use of extremely short echo spacing to reduce the susceptibility-caused geometric distortion, with a readout segmentation factor of 5, diffusion weighting in three orthogonal directions, -values of 0 and , resolution of , , and a parallel factor of 2. An additional set of diffusion tensor images (DTI) were acquired using single-shot EPI with diffusion encoding in 30 directions, -value of , TE/TR/95/4600 ms, resolution of , and a parallel imaging factor of 2. The 3-D T1-weighted dynamic contrast-enhanced (DCE) image volumes after a single dose of Gd-DTPA were acquired using a gradient echo pulse sequence with 60 dynamic phases, temporal resolution, and a voxel size of in the sagittal plane. Prior to contrast injection, 3-D gradient echo images with multiflip angles of 3, 7, 12, and 16 deg, and were acquired for quantification of T1.
The 11 patients also received PET/CT imaging on a Siemens Biograph TruePoint TrueV scanner. The average spatial resolution of the scanner is 4.4 mm FWHM.26 After intravenous injection of of 11C-MET, a 30-min dynamic 3-D mode acquisition was started. Images were reconstructed using an iterative ordered-subset expectation maximization algorithm (4 iterations, 21 subsets) with a 3-mm Gaussian filter utilizing an ultra-low-dose CT (effective mAs 30, kV 130, pitch 1.0, slice thickness 3.0 mm). Summed PET image data between 10 and 30 min were used for further evaluation.
The patients in the second dataset had pre-RT clinical MRI on a 1.5T scanner (Signa, GE). Conventional post-Gd T1-weighted images were acquired by a 2-D spin echo pulse sequence with , in-plane resolution of , 6 mm of slice thickness, and 1.5 mm of slice gap. T2-FLAIR images were required by a 2-D spin echo sequence with , in-plane resolution of , 6 mm of slice thickness, and 1.5 mm of slice gap.
PET scans of the 10 patients were performed on a Siemens ECAT EXACT HR+ whole body PET tomograph, which had an axial resolution of 4.3 mm full-width at half-maximum (FWHM) at the center of the field-of-view, which decreases to 8.3 mm FWHM at a radial distance of 20 cm.27 Following intravenous injection of of 11C-MET, a dynamic 30-min acquisition of the head was obtained in a 3-D mode. Attenuation correction was based on a transmission scan using three Ge-68 rod sources. The emission data were reconstructed iteratively using an all-pass filter with four iterations and 16 subsets.
2.2.3. Image parameter quantification
For the first dataset, the quantitative T1 maps (qT1) were estimated from multiflip angle T1-weighted images for DCE quantification. The fractional plasma volume and transfer constant () maps were derived from T1-weighted DCE MRI using the modified Tofts model implemented in-house.28 The fractional plasma volume maps were converted to CBV maps. Apparent diffusion coefficient (ADC) maps were derived from RESOLVE diffusion weighted imaging (DWI) with -values of 0 and . Fractional anisotropy (FA) images were quantified from DTI with 30-direction diffusion encoding and -values of 0 and . MET images were reconstructed using accumulated activity frames between 10 and 30 min after the agent injection.
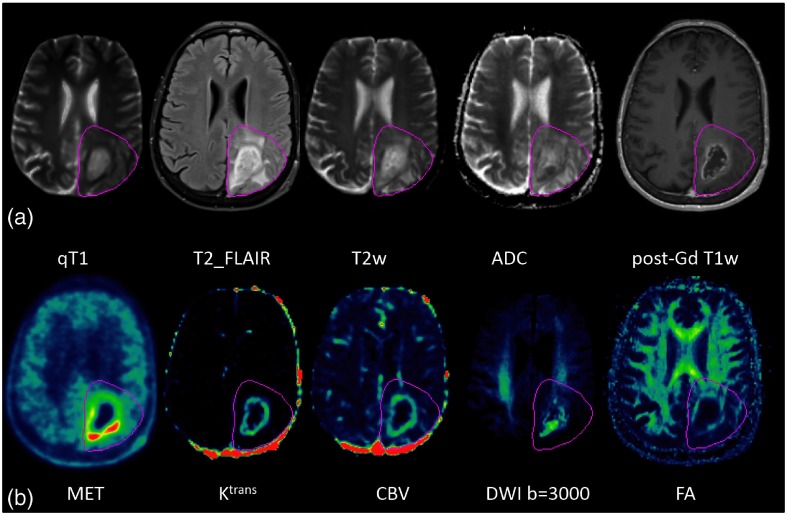
To discover image signatures and habitats in GBM, we included 10 image parameters: T2w-FLAIR, quantitative T1 (qT1), ADC, T2w (DWI with ), FA, DWI with , CBV, , MET PET, and post-Gd T1w images. The post-Gd T1 images, after being reformatted to the axial plane with a resolution of , were used as the target for registration of all images by rigid body transformation and using in-house functional imaging analysis tools. The CT acquired from PET/CT was utilized to drive image registration of MET PET with MRI.29 Examples of 10 images are shown in Fig. 2. Superpixels were created within the volume of the T2-FLAIR abnormality on the post-Gd T1w images for each tumor with a median volume of .
Fig. 2.
Ten parametric images of a patient. (a) From left, quantitative T1 (qT1), T2w-FLAIR, T2w, ADC, and post-Gd T1w images; (b) from left, MET, , CBV, DWI with , and FA images. Dark pink contours: FLAIR abnormality volume.
To test generalizability of the processing pipeline on the second dataset, we used the low resolution () of post-Gd 2-D T1 images as the target for registration of pre-RT T2w-FLAIR and MET images. The post-Gd T1-weighted images acquired at recurrence was also registered to the pre-RT images using the same method. Binary recurrence images were created to have intensity of 100 at the voxels within the recurrence tumor contour and 0 elsewhere. The pre-RT post-Gd T1-weighted, T2w-FLAIR, and MET images, and the binary recurrence images were pulled together for the analysis. Superpixels were created similarly as what was done for the first dataset.
3. Results
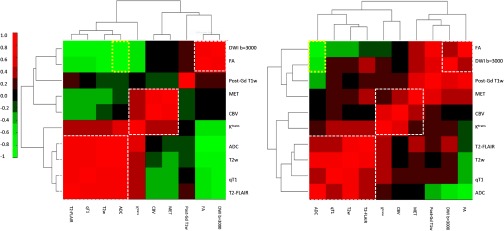
To discover the image signatures in the first dataset, the Spearman’s correlation matrix of the 10 parameters was created for each tumor and clustered according to step 3 of the processing pipeline. Two examples of the resulting correlation matrices are shown in Fig. 3. Nine parameters (except FA) had high intensity for abnormality indication. Therefore, the positive correlations between the parameters should be the signatures of interest. The correlated parameters indicate that the parameters are spatially overlapping in the image space. Three major clusters with positive correlations were identified close to the diagonal line in the heatmaps (Fig. 3): the first one included T2-FLAIR, qT1, T2w, and ADC (named as “T2_FLAIR” cluster), the second one contained MET, CBV, and (called as “MET” cluster), and the third one consisted of DWI and FA (“DWI ” cluster). The post-Gd T1w was consistently located between the “MET” and “DWI ” clusters but the correlations with the two clusters varied from tumor-to-tumor. Note that the order of the 10 parameters in the clustered matrix was not exactly the same from tumor-to-tumor, depending upon the correlation coefficients; however, the parameter members in each of the three major clusters were consistently the same. FA that had low intensity for abnormality indication showed a strong negative correlation with ADC, qT1, T2w, and T2-FLAIR across the tumors (Fig. 3). ADC showed a complex correlation pattern because both high and low intensities indicated abnormalities (edema and high cellularity, respectively). ADC was positively correlated with qT1, T2w, and T2-FLAIR, and negatively with FA and DWI (see yellow boxes in Fig. 3). showed the borderline correlation with the parameter members in the “T2-FLAIR” cluster. Note that in case (b), there were more positive correlations between the parameter members from different clusters but less such positive correlations in case (a), suggesting more spatial overlap between the parameters in the image space in case (b) than case (a). Also, the location of the post-Gd T1w in the clustergram of the correlation matrix was the same in cases (a) and (b), but the post-Gd T1w was not correlated with any parameters in case (a) but correlated with three parameters (FA, DWI , and MET) in case (b).
Fig. 3.
The clustergrams of the correlation matrices of the 10 image parameters of two patients are displayed in the heatmap. Three major clusters are revealed from the analysis. Even though the orders of the 10 parameters are different in the two patients, the parameters in each of the clusters are the same between patients.
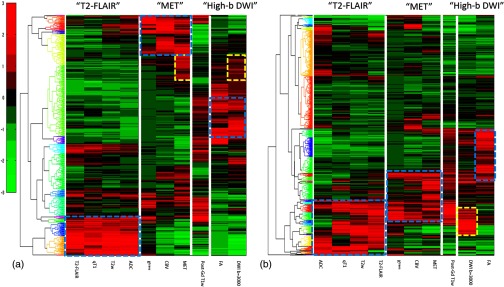
Next, we analyzed the superpixel-parameter matrix. The multimodality images exhibited a wide range of intensities. To pool multimodality image parameters and all superpixels together, we needed to standardize the parameter values. We scaled the intensities of each parameter to have zero mean and one standard deviation. Then, the superpixel-parameter matrix of each tumor was analyzed as following: (1) superpixels were clustered using the hierarchical clustering, (2) the parameter order was sorted according to the order that was found in the clustergram of the correlation matrix, and (3) the resulting matrix was displayed in the heatmap: examples shown in Fig. 4. This step of the analysis created the superpixel signatures of each tumor. The superpixel-parameter signature maps revealed three large categories of the superpixels: (1) the superpixels had high intensities distinctly within one major cluster; (2) the superpixels had high intensities in more than one major cluster, and (3) the superpixels had no high intensity in any major clusters. In our test case, the situation is unique for which 9 of the 10 parameters (except FA) had high intensities for abnormality indication. Therefore, analysis of the high intensity clusters was sufficient in our case. FA was strongly negatively correlated with the parameter members in the “T2-FLAIR” cluster and did not provide much additional information, which was ignored for the time being. In the example case shown in Fig. 4(a), the three major clusters found in the clustergram of the correlation matrix were represented by largely distinct superpixels. There was a subgroup of superpixels that had high intensities in “MET” but was modestly high in “DWI ” (yellow dashed box). In the case shown in Fig. 4(b), the three major clusters were partially overlapped among the superpixels. Also, although the parameters of DWI and FA had a modest correlation coefficient , the superpixels that had high intensity in “DWI ” were not corresponding to the ones that were high in FA.
Fig. 4.
The clustergrams of the superpixel-parameter matrices of the two GBMs in Fig. 3 displayed in the heatmaps. In case (a), the three major signature clusters have little overlap among the superpixels (cyan boxes). There was a subgroup of the superpixels having high intensity in the MET and modest-high intensity in the DWI with (yellow boxes). In case (b), the three major signature clusters had partial overlaps among the superpixels (cyan boxes). Few superpixels have high intensities in both FA and DWI with , consisting with the modest correlation coefficient () in Fig. 3(b) (yellow box).
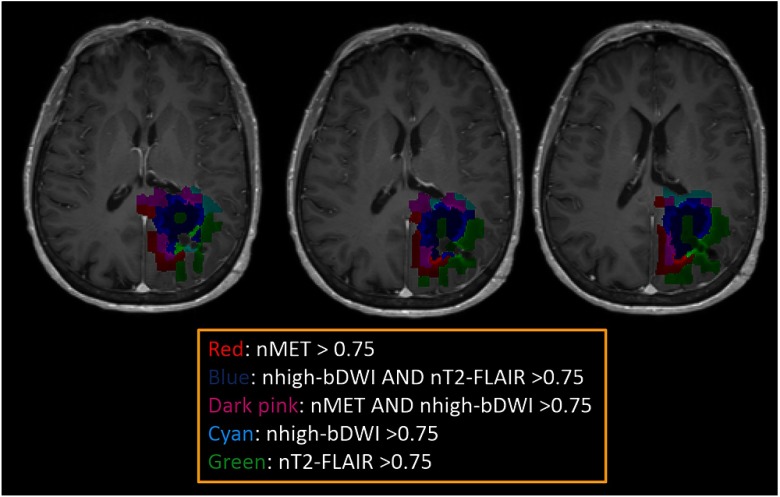
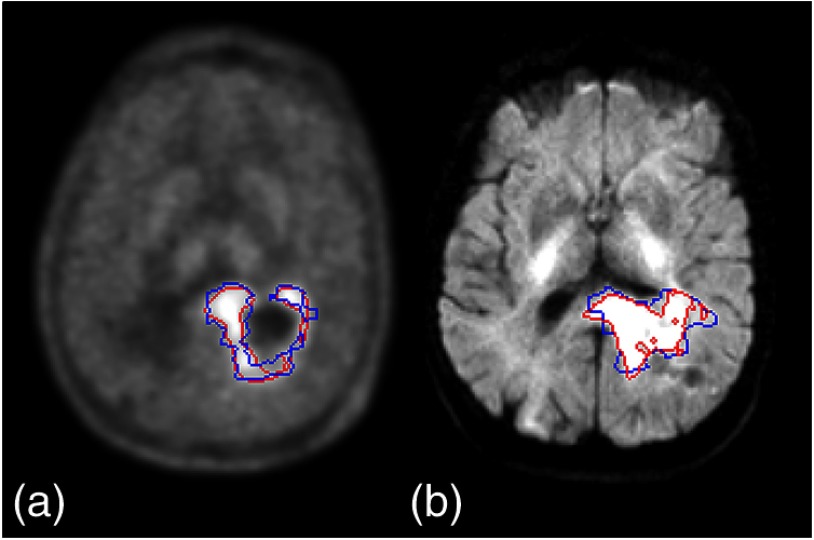
The tumor habitats can be created from the superpixel-parameter signatures by mapping and marking the superpixels belonging to the first two categories in the image space. To create major tumor habitats, a parent was chosen in each major cluster based upon if the parameter had the strongest correlation coefficients with all other members, was significantly related to tumor progression, or was a commonly used parameter in the clinic. T2-FLAIR was assigned to be the parent for cluster 1, MET was assigned to be the parent in cluster 2, and DWI for cluster 3. A cutoff threshold of the normalized intensities of “MET,” “DWI ,” and “T2-FLAIR” that defined the major signatures in Fig. 4 were used to create habitats. An example is shown Fig. 5. The tumor habitats mapped from DWI named as the “hypercellular” habitat (blue) is adjacent and peripheral to the center habitat mapped by high T2-FLAIR (green, necrotic core). The habitat mapped by both “hypercelluar” and high “MET” (dark pink) is adjacent and peripheral to the “hypercellular” habitat (blue), and is adjacent to the habitat marked by high MET only (red that is more periphery). The habitat by high “DWI ” only (cyan) is located in the periphery of the FLAIR abnormality volume. The habitat mapped by high “T2-FLAIR” only (green) makes up both the core of the tumor and the periphery. Comparing the “MET” habitat created from this analysis to the subvolume generated by a threshold of 1.5 based upon the ratio of MET intensities of tumor regions to cerebellum used in a previous study,12 we had a Dice coefficient of 78% between the two volumes. Similarly, comparing the “hypercellular” habitat to the subvolume generated by a threshold of the mean intensity plus two standard divisions in the contralateral normal tissue region of the DWI with in a previous study,11 we had a Dice coefficient of 72% between the two volumes. Examples of slices of the case in Fig. 5 are shown in Fig. 6. Note that the major discrepancies between the current methods and previous methods seem to be due to the superpixel size.
Fig. 5.
Tumor habitats color-coded and overlaid on post-Gd T1w images for the case shown in Figs. 2, 3(b), and 4(b). Red color represents the superpixels having normalized intensities of ; blue depicts the ones having normalized intensities of DWI with and T2-FLAIR ; dark pink shows superpixels having normalized intensities of MET and DWI with ; cyan denotes superpixels having DWI with , and green presents superpixels having normalized intensities of T2-FLAIR .
Fig. 6.
The habitat volumes (blue color) of (a) “MET” and (b) “hypercellularity” compared to the subvolumes (red color) defined by previous methods. The major differences seem to be due to the superpixels.
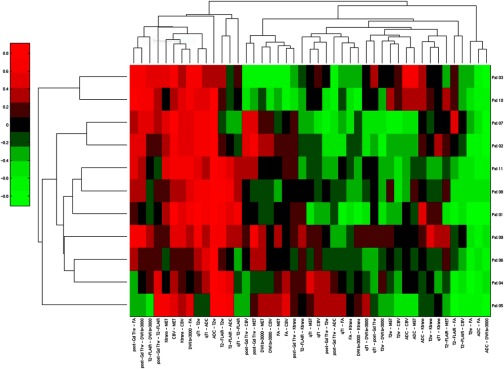
Finally, to discover the subtypes of GBM, which could be associated with different progression or outcome patterns, all image parameter pairs of the correlation coefficients from all patients were pooled together and analyzed by hierarchical clustering, see Fig. 7. The clustergram shows that the image signatures vary from the patient at the top row to the one at the bottom row. Note that there were redundant pairs of image parameters, and pairs that were uniform across patients. For example, the three pairs in first three right columns had uniform correlation coefficients across patients. If the patients had different outcomes or progression time and patterns, then the three pairs could not differentiate progression patterns or outcomes.
Fig. 7.
Clustergram of the matrix with the Spearman’s correlation coefficients of all parameter pairs from all patients in a heatmap for discovery of the subtypes of GBM.
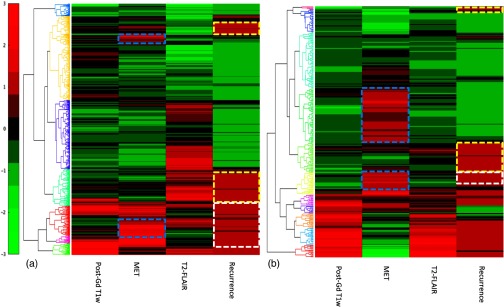
To test the generalizability of the processing pipeline, the superpixel-parameter matrix, including the recurrence binary maps, was created for each case in the second dataset. We did not find unexpected results from the second dataset due to different acquisitions compared to the first one. Two examples are shown in Fig. 8. Note that there were distinct patterns of Gd enhancement, MET uptake, and T2-FLAIR abnormality in two cases. Both cases had MET uptake occurred in the superpixels without Gd enhancement. The clustergrams of the superpixel matrices also revealed the association or disassociation of “MET signatures” and “Gd enhancement signatures” with recurrence. In the first case, there were of recurrence occurred in the superpixels with MET uptake and 50% with no spatial coincidence of either Gd enhancement or MET uptake. In the second case, there was a small portion of recurrence occurred in the superpixels with MET uptake. There were of recurrence occurred in the superpixels that had neither Gd enhancement, nor MET uptake, suggesting other image-modality parameters are needed to further characterize the heterogeneity of GBM. Figure 9 shows the clustergram of Spearman’s correlation coefficients of tested image parameters and recurrence maps of the 10 patients from the second dataset. The clustergram shows that in five patients, the superpixels that were coincident with recurrent locations were correlated with the locations of high MET uptake pretreatment but not in other five patients, suggesting that the tool that was developed in the study can be used to investigate the recurrent patters with imaging habitats.
Fig. 8.
Clustergrams of the superpixel-parameter matrices of two patients with GBM from the second dataset. In the first case (a), only of the superpixels with MET uptake were Gd enhanced. In the second case (b), the majority of the superpixels with MET uptake were not enhanced by Gd. Cyan boxes mark the superpixels with MET uptake but not enhanced by Gd. Note that recurrence occurred in the superpixels with (white box) and without (yellow box) MET uptake pre-RT.
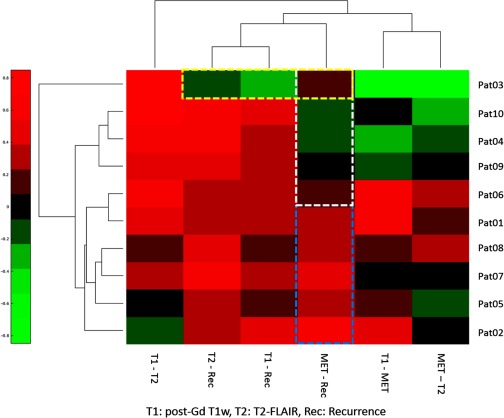
Fig. 9.
Clustergram of the matrix of the Spearman’s correlation coefficients of image parameters and recurrence maps of the 10 patients from the second dataset. Note that five patients had modest to high correlation () between recurrence and MET uptake (cyan box) and the other five patients did not (white box). One patient had no positive correlations between recurrence and tested image parameters.
4. Discussion
We have developed a framework of a processing pipeline for discovery of tumor image-defined signatures and habitats. The framework consists of (1) assigning multi-image parameters on superpixels, (2) discovering major image signatures in each tumor, (3) clustering the superpixels based upon the major signatures, and (4) creating the tumor habitats from the image signatures. For a proof-of-concept, we applied the processing pipeline to 11 patients with GBM. We discovered the three major image signatures from 10 image parameters and created five major habitats. We further tested the processing pipeline in the second dataset for generalizability and the clinical association. We show that the association of recurrence patterns with image parameter-defined signatures can be revealed. The clinical meanings of the GBM habitats, and whether one or two habitats are strongly associated with clinical tumor progression and/or “relapse” genomic profile need to be investigated further in clinical trials and genomic sequencing of image-guided biopsy tissue in future studies.
Our framework for discovery of the image-define signatures and habitats can be further tailored to specific tumor types and a set of images. Using superpixels reduces the redundancy in the original pixels and improves the image processing efficiency. Large size and compactness allow more parameter variations within a single superpixel. The size of the superpixel should be large enough to minimize the impact of potential spatial misalignment of images but small enough to preserve image details of interest. In addition, for a set of images that have different intrinsic resolutions, e.g., PET versus MRI, how to choose the basic superpixel size needs to be further investigated. The superpixel can be defined on a single image or an image vector. The trade-off between the two is that the latter can capture variations in the image parameters of interest better but also may be affected by noise and artifact. All these tradeoffs need to be investigated in specific tumor types and image types.
Our signature discovery process in GBM as a proof-of-concept seems to work well. Our strategy is to discover the major signatures first in the covariance or correlation matrix of the whole tumor. The process at this level neglects that there may be a subgroup of superpixels that have different signatures from the ones discovered by analyzing the whole tumor. Such subgroups of superpixels can be discovered in the clustergram of the superpixel-parameter matrix and mapped to the image space as “subhabitats” for further clinical studies. In this study, we learned that the major signatures would be close to the diagonal line in the covariance or correlation matrix if all abnormal image parameters have high intensities. These types of the patterns are more easily learned than a pattern of mixed positive and negative correlations. Prior knowledge of the parameters could be used to organize and transform all abnormal parameters to have high intensity. For example, FA can be transformed to 1-FA that shows positive correlations with qT1, T2w, T2-FLAIR, and ADC. If there is no prior knowledge to indicate high or low intensity for abnormality indication, a two-step analysis can be used. The first step is to determine whether the abnormal parameter of interest is negatively correlated with the parameters with high intensity. After that, the parameter can be transformed accordingly for clustering analysis. It is worthwhile to point out that the signature discovery process can reveal the degree of redundancy in the image parameters. The latter can be eliminated from the analysis or even acquisition. In our analysis, we also noted that the surgical cavity in the cases with gross total resection can affect the correlation of T2-FLAIR with other parameters. Excluding the surgical cavity leads to more consistent clustergrams.
The methods that integrate all image parameters of the superpixels into a single matrix need to be further developed and evaluated. In our superpixel-parameter matrix analysis, intensity standardization was used to deal with the wide range of the values of the multimodality images and to transform the parameters to the same range. We scaled all parameters to have the same mean and standard deviation. Our data parameters are not perfectly normally distributed, which did not appear to affect the results. How much deviation of a parameter distribution from the normal distribution would affect the results needs to be tested further. The histogram matching that does not require a normal distribution of a parameter could be investigated for the intensity standardization.30 However, image artifacts with large intensities affect the results, which should be removed from the analysis. To create tumor habitats, a cutoff threshold was used to create the binary habitat in this study for a proof-of-concept. The cutoff threshold was chosen empirically from the clustergrams in Fig. 4, which needs to be evaluated further. A preliminary evaluation shows the high similarity between the habitat volumes and the subvolumes created using previous methods.11,12 Further refining the superpixels could improve the habitat definition. The robustness of the use of a single threshold to define the habitats after standardizing the data intensities needs to be further investigated. Histopathology is the gold standard to evaluate and establish a cutoff threshold, which is often not available. Clinical outcome data, e.g., progression location, could be also used to establish a practical cutoff threshold for clinical usage. A recurrence probability map of a habitat can also be created. All these need to be further investigated in future studies.
While testing two small datasets with MET PET, we found only 30% to 50% of patients in whom the superpixels with MET uptake in the FLAIR abnormality were strongly correlated with the contrast enhancement, indicating that the contrast enhancement is not sufficient to describe GBMs, and other imaging modalities are needed. We also learn that although providing additional metabolic information in GBM, MET uptake does not define all habitats and is not correlated with recurrent sites in all patients. Given the small number of patients, it is premature to make any further clinical conclusions. However, our tools can be a means to perform this kind of analysis to discover what image habitats are associated with recurrence. Finally, our tools have an extent of tolerance on image acquisition differences, which could, at least partially, attribute to the use of superpixels.
In this study, we focused on imaging characteristics to create clustergrams from all parameter pairs of correlation coefficients and all patients. However, other data can be entered into the matrix for the analysis, e.g., the size of certain habitats. Also, molecular biomarkers—e.g., MGMT methylation and IDH mutation status—and genomic data can be added into the matrix for analysis.
In conclusion, we have developed a framework of the processing pipeline to discover tumor image-defined signatures and then create tumor habitats. Our approach aimed to link the image signatures to habitats, which has not been done before. Further development and validation will be conducted in a cohort of patients with histopathology, genomic sequencing, and clinical outcomes.
Acknowledgments
This work was supported in part by the NIH 1U01CA183848.
Biographies
Daekeun You is a senior software developer in the Department of Radiation Oncology, University of Michigan. He has been working on various research projects in fields of quantitative imaging, information retrieval, and pattern recognition. He received his PhD in computer science and engineering from the State University of New York at Buffalo in 2011.
Yue Cao, PhD, FAAPM, professor of Departments of Radiation Oncology, Radiology and Biomedical Engineering of the University of Michigan. Her research interests are development of quantitative imaging and analysis methods for tumor and normal tissue response assessment. She is PI and Co-PI on NIH RO1 and UO1 grants. She has published more than 140 papers and book chapters.
Biographies for the other authors are not available.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Keles G. E., et al. , “Volume of residual disease as a predictor of outcome in adult patients with recurrent supratentorial glioblastomas multiforme who are undergoing chemotherapy,” J. Neurosurg. 100, 41–46 (2004). 10.3171/jns.2004.100.1.0041 [DOI] [PubMed] [Google Scholar]
- 2.Stupp R., et al. , “Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma,” N. Engl. J. Med. 352, 987–996 (2005). 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 3.Cao Y., et al. , “The extent and severity of vascular leakage as evidence of tumor aggressiveness in high-grade gliomas,” Cancer Res. 66, 8912–8917 (2006). 10.1158/0008-5472.CAN-05-4328 [DOI] [PubMed] [Google Scholar]
- 4.Cao Y., et al. , “Physiologic and metabolic magnetic resonance imaging in gliomas,” J. Clin. Oncol. 24, 1228–1235 (2006). 10.1200/JCO.2005.04.7233 [DOI] [PubMed] [Google Scholar]
- 5.Cao Y., et al. , “Clinical investigation survival prediction in high-grade gliomas by MRI perfusion before and during early stage of RT,” Int. J. Radiat. Oncol. Biol. Phys. 64, 876–885 (2006). 10.1016/j.ijrobp.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 6.Chenevert T. L., et al. , “Diffusion magnetic resonance imaging: an early surrogate marker of therapeutic efficacy in brain tumors,” J. Natl. Cancer Inst. 92, 2029–2036 (2000). 10.1093/jnci/92.24.2029 [DOI] [PubMed] [Google Scholar]
- 7.Graves E. E., et al. , “A preliminary study of the prognostic value of proton magnetic resonance spectroscopic imaging in gamma knife radiosurgery of recurrent malignant gliomas,” Neurosurgery 46, 319–326 (2000). 10.1097/00006123-200002000-00011 [DOI] [PubMed] [Google Scholar]
- 8.Grosu A. L., et al. , “Positron emission tomography for radiation treatment planning,” Strahlenther. Onkol. 181, 483–499 (2005). 10.1007/s00066-005-1422-7 [DOI] [PubMed] [Google Scholar]
- 9.Hamstra D. A., et al. , “Functional diffusion map as an early imaging biomarker for high-grade glioma: correlation with conventional radiologic response and overall survival,” J. Clin. Oncol. 26, 3387–3394 (2008). 10.1200/JCO.2007.15.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law M., et al. , “Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging,” Radiology 247, 490–498 (2008). 10.1148/radiol.2472070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pramanik P. P., et al. , “Hypercellularity components of glioblastoma identified by high b-value diffusion-weighted imaging,” Int. J. Radiat. Oncol. Biol. Phys. 92, 811–819 (2015). 10.1016/j.ijrobp.2015.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsien C. I., et al. , “Concurrent temozolomide and dose-escalated intensity-modulated radiation therapy in newly diagnosed glioblastoma,” Clin. Cancer Res. 18, 273–279 (2012). 10.1158/1078-0432.CCR-11-2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee I. H., et al. , “Association of 11C-methionine PET uptake with site of failure after concurrent temozolomide and radiation for primary glioblastoma multiforme,” Int. J. Radiat. Oncol. Biol. Phys. 73, 479–485 (2009). 10.1016/j.ijrobp.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahl D. R., et al. , “Combined imaging of elevated CBV and hypercellularity in glioblastoma to inform management and intensify treatment of resistant tumor subvolumes,” Int. J. Radiat. Oncol. Biol. Phys. 96, S182–S183 (2016). 10.1016/j.ijrobp.2016.06.456 [DOI] [Google Scholar]
- 15.Eisen M. B., et al. , “Cluster analysis and display of genome-wide expression patterns,” Proc. Natl. Acad. Sci. U. S. A. 95, 14863–14868 (1998). 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spellman P. T., et al. , “Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization,” Mol. Biol. Cell 9, 3273–3297 (1998). 10.1091/mbc.9.12.3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamayo P., et al. , “Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation,” Proc. Natl. Acad. Sci. U. S. A. 96, 2907–2912 (1999). 10.1073/pnas.96.6.2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov L. B., et al. , “Signatures of mutational processes in human cancer,” Nature 500, 415–421 (2013). 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillies R. J., Kinahan P. E., Hricak H., “Radiomics: images are more than pictures, they are data,” Radiology 278, 563–577 (2016). 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalpathy-Cramer J., et al. , “Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features,” Tomography 2, 430–437 (2016). 10.18383/j.tom.2016.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aerts H. J., et al. , “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat. Commun. 5, 4006 (2014). 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M., et al. , “Superpixel segmentation: a benchmark,” Signal Process. Image Commun. 56, 28–39 (2017). 10.1016/j.image.2017.04.007 [DOI] [Google Scholar]
- 23.Achanta R., et al. , “SLIC superpixels compared to state-of-the-art superpixel methods,” IEEE Trans. Pattern Anal. Mach. Intell. 34, 2274–2282 (2012). 10.1109/TPAMI.2012.120 [DOI] [PubMed] [Google Scholar]
- 24.Bar-Joseph Z., Gifford D. K., Jaakkola T. S., “Fast optimal leaf ordering for hierarchical clustering,” Bioinformatics 17(Suppl. 1), S22–S29 (2001). 10.1093/bioinformatics/17.suppl_1.S22 [DOI] [PubMed] [Google Scholar]
- 25.MathWorks, “R2017b Documentation,” https://www.mathworks.com/help/bioinfo/ref/clustergram.html.
- 26.Jakoby B. W., et al. , “Performance characteristics of a new LSO PET/CT scanner with extended axial field-of-view and PSF reconstruction,” IEEE Trans. Nucl. Sci. 56, 633–639 (2009). 10.1109/TNS.2009.2015764 [DOI] [Google Scholar]
- 27.Brix G., et al. , “Performance evaluation of a whole-body PET scanner using the NEMA protocol,” J. Nucl. Med. 38, 1614–1623 (1997). [PubMed] [Google Scholar]
- 28.Tofts P. S., et al. , “Estimating kinetic parameters from dynamic contrast-enhanced t1-weighted MRI of a diffusable tracer: standardized quantities and symbols,” J. Magn. Reson. Imaging 10, 223–232 (1999). 10.1002/(ISSN)1522-2586 [DOI] [PubMed] [Google Scholar]
- 29.Wienhard K., et al. , “The ECAT EXACT HR: performance of a new high resolution positron scanner,” J. Comput. Assist. Tomogr. 18, 110–118 (1994). 10.1097/00004728-199401000-00023 [DOI] [PubMed] [Google Scholar]
- 30.Nyúl L. G., Udupa J. K., Zhang X., “New variants of a method of MRI scale standardization,” IEEE Trans. Med. Imaging 19, 143–150 (2000). 10.1109/42.836373 [DOI] [PubMed] [Google Scholar]