Abstract
With >100 common variants associated with schizophrenia risk, establishing their biological significance is a priority. We sought to establish cognitive effects of risk variants at loci implicated in synaptic transmission by (1) identifying GWAS schizophrenia variants whose associated gene function is related to synaptic transmission, and (2) testing for association between these and measures of neurocognitive function. We selected variants, reported in the largest GWAS to date, associated with genes involved in synaptic transmission. Associations between genotype and cognitive test score were analyzed in a discovery sample (988 Irish participants, including 798 with psychosis), and replication samples (528 UK patients with schizophrenia/schizoaffective disorder; 921 German participants including 362 patients with schizophrenia). Three loci showed significant associations with neuropsychological performance in the discovery samples. This included an association between the rs2007044 (risk allele G) within CACNA1C and poorer working memory performance (increased errors B (95% CI)=0.635–4.535, p=0.012), an effect driven mainly by the psychosis groups. In an fMRI analysis of working memory performance (n=84 healthy participants, a subset of the discovery sample), we further found evidence that the same CACNA1C allele was associated with decreased functional connectivity between the right dorsolateral prefrontal cortex and right superior occipital gyrus/cuneus and anterior cingulate cortex. In conclusion, these data provide evidence to suggest that the CACNA1C risk variant rs2007044 is associated with poorer memory function that may result from risk carriers’ difficulty with top-down initiated responses caused by dysconnectivity between the right DLPFC and several cortical regions.
Introduction
Predictions that increasing sample sizes would also lead to an increase in genome-wide significant schizophrenia (SZ) risk variants being identified (Park et al, 2010) have been supported to date. As sample sizes available for GWAS have increased, the numbers of significantly associated common variants and genes has gone from one, ZNF804A (O'Donovan et al, 2008), to a handful (The Schizophrenia Psychiatric GWAS Consortium, 2011) to over 108 loci now being implicated (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). As the purpose of gene discovery is to identify the biological pathways that increase risk and are capable of being targeted by treatments, moving from discovering to establishing functional impact is a priority for our field.
Although this is often thought of at the molecular and cellular levels, establishing the functional impact of genes and gene sets at other levels of analysis (ie, neural, cognitive, and behavioral) is also likely to be important in providing insights for new treatment discovery. We and others have previously sought to establish the cognitive effects of polygenic risk in pathways defined either by biological processes, eg, variants within the cell adhesion molecule pathway (Hargreaves et al, 2013), or by gene sets functionally related to identified SZ-risk genes (eg, MIR137, ZNF804A) (Cosgrove et al, 2017; Nicodemus et al, 2014). To date, and to our knowledge, however, no study has systemically investigated the cognitive effects of SZ variants for synaptic transmission despite the fact that alterations in this process (and neurotransmitter systems, eg, dopamine) have long been held as a key etiological hypothesis for both SZ risk and deficits in cognition associated with the disorder (Andreasen, 1995; Pocklington et al, 2014; Sarter et al, 2007). Looking at the effect of SZ-associated variants on cognition may help identify specific neurobiological processes involved, so as to allow this aspect of disability to be more effectively targeted.
The purpose of the present study was to undertake a systematic analysis of the cognitive effects of SZ risk-associated variants identified by the Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC-SZ) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) in genes related to synaptic transmission using a novel methodology. We did this by (1) cross-referencing PGC-SZ-identified variants whose associated gene function was bioinformatically identified as related to synaptic transmission using OMIM and Gene Ontology, and (2) comparing the performance of carriers and noncarriers of these variants on cognitive functions typically impaired in SZ. Our hypothesis was that the identified risk variants at these gene loci would be associated with poorer performance on measures of cognition.
Materials and methods
Neuropsychological Sample Characteristics
Cognitive performance was assessed in 939 cases and 330 healthy participants (769 males, 500 females). Cases consisted of clinically stable patients with either a ‘narrow sense psychosis’ diagnosis of SZ or schizoaffective disorder (SZA) (n=676) or a ‘broader sense psychosis’ that, in addition to our SZ/SZA sample, included patients with a diagnosis of bipolar disorder with psychotic features, major depressive disorder with psychotic features, or psychosis not otherwise specified (n=263), based on Structured Clinical Interview for DSM-IV Axis I (First et al, 2002).
Two replication samples were also available. Samples from Cardiff consisted of 528 European Caucasian individuals with SZ or SZA who were recruited from community mental health teams in Wales and England. The German replication sample consisted of 362 clinically stable patients with a DSM-IV diagnosis of SZ and 559 healthy controls, all genotyped as part of a previous study (O'Donovan et al, 2008). A full description of the discovery and replication samples can be found in Supplementary Appendix 1.
Demographic variables available for all samples included age, gender, and years of education. For patients, both clinical symptom severity (measured using the Scale for the Assessment of Positive Symptoms (SAPS) and the Scale for the Assessment of Negative Symptoms (SANS)) and medication dosage in terms of chlorpromazine equivalents (CPZ) were also available, as previously described (Walters et al, 2010).
fMRI Spatial Working Memory Task Sample Characteristics
A subgroup of the healthy participants from the Irish discovery sample (n=103, all right handed) had also undergone functional imaging during performance of an fMRI spatial working memory (SWM) task. This task, and the acquisition parameters used, has been extensively used by us previously (Rose et al, 2012a, b, 2013). This task required participants to remember the spatial location of a stimulus (either one white dot or three white dots) after a 3 s delay. See Supplementary Appendix 1 for a full description.
Candidate SNP Selection
A novel approach was taken to gene selection in this study. All genes located in the genome-wide significant regions (Supplementary Table S1a) in the largest SZ GWAS to date (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) were assigned a function using the OMIM (OMIM, 2014) and Uniprot (UniProt Consortium, 2014) catalogs. Genes associated with the processes of neurotransmission, neurotransmitter secretion, or transduction were identified by the lead author. Following this, 28 genes functioning in synaptic transmission were prioritized (Supplementary Table S1b). We examined the association signals at the GWAS loci containing these genes, initially by visual inspection of the supplementary regional GWAS association plots from the PGC-SZ (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). For 10 of the 28 genes, the association signal at the GWAS locus mapped to within the boundaries of a synaptic transmission gene. Nine of the index SNPs were intronic with synaptic transmission genes and the tenth SNP was rs2514218 upstream of DRD2. For the remaining genes, we checked for cis effects of these SNPs on genes using the Braineac (Ramasamy et al, 2014) and GTEx (https://www.gtexportal.org/, The GTEx Consortium, 2013) databases where p-values were corrected for the number of brain tissues tested (n=10 for Braineac, n=13 for GTEx). For RIMS1, rs1339227 was associated with altered gene expression in the thalamus (Braineac, corrected p=0.043). Overall, this exercise shortlisted 11 of the 28 genes where the genome-wide significant association signal was at or close to a synaptic transmission gene or the index SNP had a cis effect on gene expression (Supplementary Table S1c). For these loci we identified the most significant GWAS SNP (the index SNP) as the variant for use in our cognition analyses. This gene selection process is illustrated in Supplementary Figure S1. In the case of CACNA1C multiple GWAS signals relating to two LD blocks were identified; for this reason two SNPS were included in the analysis, representing the strongest signal for each LD block. Finally, to bioinformatically confirm functional identity, this list of genes was entered into GO, http://www.geneontology.org (The Gene Ontology Consortium, 2015), and found to have >98-fold enrichment in GO term ‘chemical synaptic transmission, postsynaptic’ (GO: 0099565). Based on this approach the following variants were identified: rs2007044 and rs2239063 (CACNA1C), rs7893279 (CACNB2), rs8042374 (CHRNA3), rs10520163 (CLCN3), rs2514218 (DRD2), rs9922678 (GRIN2A), rs12704290 (GRM3), rs1501357 (HCN1), rs1339227 (RIMS1), and rs4523957 (SRR).
Neuropsychological Assessment
The second stage in the study was to undertake a cognitive analysis of the variants identified above, seeking replication of significant findings in independent samples. Participants completed a full neuropsychological battery of measures indexing the cognitive deficits typically reported in SZ (with five cognitive domains—general cognitive function, episodic and working memory, attention, and social cognition). Selected subtests (Vocabulary, Similarities, Block Design, and Matrix Reasoning) of the Wechsler Adult Intelligence Scale (WAIS III) (Wechsler, 1997a) were used to measure general cognitive function. The Logical Memory and Faces subtests from the Wechsler Memory Scale (WMS III) (Wechsler, 1997b) were used to assess verbal and visual memory recall. Working memory was assessed using the SWM task taken from the Cambridge Automated Neuropsychological Test Battery (CANTAB) (Cambridge Cognition, 2016) and the Letter–Number Sequencing (LNS) subtest from WAIS III. Attentional control was assessed using the Continuous Performance Task (CPT) (Cornblatt et al, 1988) and the Sustained Attention to Response Task (SART) (Robertson et al, 1997). Social cognitive function was assessed using three measures of social cognition: two Theory of Mind (ToM) tests and one attributional style test. The ToM tests used were the Reading the Mind in the Eyes test (Eyes) (Baron-Cohen et al, 2001) and the Hinting Task (Hint) (Corcoran et al, 1995). Attributional style was measured using the Internal, Personal, Situational Attributions Questionnaire (IPSAQ) (Kinderman and Bentall, 1996).
All individuals in the Cardiff sample were assessed using the MATRICS Consensus Cognitive Battery (Nuechterlein and Green, 2006). This includes seven cognitive domains: speed of processing, attention/vigilance, verbal and nonverbal working memory, verbal and visual memory, reasoning, and problem solving. Of note, the social cognition domain included in the MATRICS was not a measure of ToM (unlike in the Irish samples) but rather an index of emotional self-regulation.
In the German sample, IQ was measured using the German version of the WAIS Revised (Wechsler, 1991), and memory and attention was assessed using subtests from the German WMS Revised (Härting et al, 2000), the n-back (Callicott et al, 2000; Egan et al, 2001), and the Continuous Performance Test, 3–7 Version (Nuechterlein and Asarnow, 2004), respectively. These measures were selected on the basis of closely approximating the measures available in the Irish samples; unfortunately, measures of social cognition were not available for the German samples. Supplementary Table S2 provides more detailed description of neuropsychological tests used in each sample.
Genotyping
Genotyping was performed on DNA extracted from whole blood or saliva on a subset of participants who underwent neuropsychological testing. Full GWAS data were available for 988 participants (healthy participants n=190; SZ/SZA=578; other psychoses n=220). A fuller description can be found in Supplementary Appendix 1.
Statistical Analysis
Analysis of neuropsychological data
All neuropsychological analyses were carried out using SPSS 20 (IBM, 2012). To estimate risk variant effects on cognitive deficits associated with SZ, linear regression analyses were performed on the total sample (ie, irrespective of diagnosis, n=988). To test associations between genotype and cognitive test score, hierarchical multiple regression with bootstrapping was performed (1000 iterations, with replacement, to identify a bias corrected and accelerated 95% confidence interval). In each case cognitive test score was entered as a dependent variable, and age and sex were entered as predictors on the first step, followed by genotype on the second step. As this initial discovery sample analysis was carried out in order to identify any significant or nominally significant findings, a correction for multiple testing was not applied. Instead, nominally significant findings from cognitive analyses were taken forward for analysis to seek replication in two independent samples. Where replication of results was found in these independent samples, the associated SNPs were taken forward for neuroimaging analysis.
Analysis of fMRI data
Preprocessing and statistical analysis are detailed in Supplementary Appendix 1. First-level analysis included working memory (1 dot and 3 dots>baseline) and increased memory load (3 dots>1 dot) contrasts. The 1 dot and 3 dots>baseline contrast considered both the 1-dot and 3-dot conditions together (ie, all of the spatial working memory blocks) and contrasted all of these blocks against the baseline blocks (the blocks during which there was no delay between the white dot and red circle). The 3 dots>1 dot condition considered the 3-dot blocks and contrasted these against the 1-dot blocks to examine increasing blood-oxygen-level-dependent (BOLD) response associated with increasing working memory load (ie, remembering the location of 3 white dots compared with remembering the location of 1 white dot). Contrast maps were entered into a multiple regression analysis with number of risk alleles as covariate of interest. Out of 103 participants, 19 were excluded from fMRI activation analysis because of low quality or missing data (Supplementary Appendix 1). Seed voxel correlation analysis was used to examine functional connectivity using the right dorsolateral prefrontal cortex (DLPFC) as a seed. Eigenvariates extracted from this seed were entered into a first-level analysis and resulting connectivity maps entered into a multiple regression analysis. Results were examined at a p<0.001 (uncorrected) level and clusters were considered statistically significant at a p<0.05 level, family-wise error (FWE) corrected for multiple comparisons across the whole brain at the cluster level. Masks used for functional connectivity analysis were kindly provided to us by C. Esslinger and F. Paulus (Paulus et al, 2014). Functional connectivity analysis was examined using both ‘global maximum’ and ‘next local maximum’ seed selection methods to examine whether results were consistent across both methods (Bedenbender et al, 2011) (Supplementary Appendix 1). The rationale for carrying out this connectivity analysis is that genetic effects may be more penetrant at a cortical level than a behavioral level, as we and others have previously shown for SZ risk variants (Paulus et al, 2014; Rose and Donohoe, 2013).
Results
Neuropsychological Samples: Demographic and Clinical Characteristics
For the cognitive measures employed, healthy participants scored higher than patients on all measures of IQ, attention, and social cognition used in analysis except for Externalizing Bias (ANCOVA; covariates age and gender, p<0.0001; Table 1). There was a significant difference in age between the rs9922678 (GRIN2A) allele groups only (F=4.28, p<0.05; mean values GG=41.15, GA=42.44, AA=38.28). No other between-group differences were observed. Supplementary Table S4 shows means and SDs for each SNP and neuropsychological/demographic variables in the discovery sample.
Table 1. Neuropsychological Sample Participant Demographics and Subtest Scores (Irish Discovery Sample).
| Measure, mean (SD) | Healthy participants | N | Broad Diagnosis | N | F (healthy V broad) | Narrow diagnosis | N | F (healthy V narrow) |
|---|---|---|---|---|---|---|---|---|
| Age | 35.82 (12.65) | 330 | 43.00 (12.40) | 939 | 81.02*** | 42.30 (12.47) | 676 | 59.36*** |
| Male/female (%) | 44.5 : 55.5 | 330 | 66.2 : 33.8 | 939 | 49.96*** | 70.4 : 29.6 | 676 | 67.01*** |
| CPZ, mg/day | — | — | 435.97 (453.17) | 679 | — | 498.18 (482.18) | 517 | — |
| SAPS | — | — | 19.86 (19.30) | 771 | — | 21.86 (19.77) | 595 | — |
| SANS | — | — | 23.58 (19.93) | 767 | — | 26.38 (19.99) | 593 | — |
| Education years | 15.64 (1.82) | 324 | 12.95 (2.64) | 713 | 184.28*** | 12.73 (2.51) | 516 | 191.51*** |
| WTARR | 41.90 (5.96) | 329 | 33.52 (10.60) | 715 | 179.36*** | 32.77 (10.56) | 524 | 194.60*** |
| VERBIQ | 118.45 (15.80) | 326 | 92.06 (20.02) | 865 | 374.27*** | 90.36 (19.18) | 635 | 433.73*** |
| PERFIQ | 117.78 (22.90) | 323 | 91.20 (19.32) | 677 | 356.35*** | 89.83 (18.43) | 496 | 393.58*** |
| FSIQ | 119.68 (15.43) | 320 | 92.07 (19.22) | 670 | 429.87*** | 90.31 (18.12) | 490 | 515.19*** |
| LM1 | 49.09 (10.10) | 317 | 27.95 (12.35) | 918 | 597.47*** | 26.52 (12.37) | 655 | 627.06*** |
| LM2 | 31.75 (7.44) | 315 | 15.34 (8.84) | 906 | 688.13*** | 14.40 (8.68) | 648 | 732.18*** |
| FACES1 | 38.93 (4.50) | 284 | 33.79 (5.09) | 553 | 128.61*** | 33.65 (5.20) | 431 | 124.70*** |
| FACES2 | 39.56 (4.30) | 284 | 35.04 (5.28) | 547 | 85.29*** | 35.04 (5.24) | 426 | 85.14*** |
| LNS | 12.72 (3.13) | 326 | 8.16 (3.40) | 886 | 353.15*** | 7.90 (3.44) | 632 | 365.55*** |
| CANTAB SWM (errors) | 19.04 (16.45) | 274 | 43.05 (23.85) | 652 | 189.727*** | 43.86 (24.21) | 473 | 204.20*** |
| SART (reaction time) | 32.38 (90.80) | 182 | 445.29 (112.64) | 502 | 126.04*** | 443.83 (113.42) | 393 | 122.46*** |
| CPT d’Prime 2 digit | 3.82 (0.56) | 41 | 2.69 (1.21) | 498 | 19.95*** | 2.67 (1.22) | 365 | 19.79*** |
| CPT d’Prime 3 digit | 3.35 (0.75) | 41 | 3.35 (0.75) | 483 | 46.37*** | 1.91 (1.09) | 352 | 46.72*** |
| CPT d’Prime 4 digit | 2.25 (0.97) | 41 | 1.14 (0.87) | 448 | 45.76*** | 1.12 (0.83) | 327 | 48.38*** |
| Eyes | 26.47 (3.58) | 273 | 21.21 (6.21) | 469 | 101.45*** | 20.48 (6.39) | 329 | 120.43*** |
| Hint | 16.86 (1.79) | 224 | 15.56 (3.31) | 439 | 23.12*** | 15.34 (3.48) | 338 | 27.94*** |
| IPSAQ–externalizing bias | 1.52 (4.02) | 228 | 1.49 (3.85) | 410 | 0.54 | 1.39 (3.88) | 316 | 0.34 |
| IPSAQ–personalizing bias | 0.62 (0.24) | 230 | 0.52 (0.29) | 410 | 16.39*** | 0.51 (0.28) | 318 | 18.69*** |
Abbreviations: CPZ, chlorpromazine equivalents; FSIQ, Full-Scale IQ; SANS, Scale for the Assessment of Negative Symptoms; SAPS, Scale for the Assessment of Positive Symptoms.
***P<0.001.
When genotype groups were compared on demographic variables, no significant between-group differences were observed for any SNP included for either gender or years of education (p>0.05). No between-group differences were observed for diagnosis status or total positive symptom severity scores (SAPS total scores) for any SNP. For symptoms, the DRD2 rs2514218 risk genotype showed significantly higher negative symptoms (F=3.67, p<0.05; mean values TT=18.61, CT=23.78, CC=25.69). No medication dosage (measured in terms of chlorpromazine equivalents) or other genotype-related clinical differences were observed. Demographic and clinical characteristics for patients and healthy participants are presented in Table 1. A between-group comparison of these groups indicated differences between the healthy participants and both broad and narrow groups in terms of age, gender, and education. Supplementary Table S3 shows summary statistics and results comparing genotype group demographics of SNPs showing a significant effect on any neuropsychological variable. No age or sex associations were observed with rs2007044 in either the Cardiff or German sample.
Effect of Genotype on Cognition
Analyses were performed separately for each SNP on the sample as a whole (all cases and healthy participants) for each neuropsychological test (18 tests covering 5 cognitive domains). When significant results were found, this was followed up using subgroup analysis to determine the effects of diagnosis (broad psychosis group, narrow psychosis group, and healthy participants). Of 11 SNPs included, significant effects were observed in the discovery sample (cases and healthy participants) for three SNPs (Table 2 presents the three SNPs with significant results in all participants and subgroups; Supplementary Table S4 presents the means and SDs of the whole discovery sample for each SNP). For rs2007044 (located within the calcium channel subunit α1c (CACNA1C) gene), increased risk ‘G’ allele dosage was associated with increased errors made on the CANTAB SWM, explaining a proportion of test score variance in the entire sample (B=2.6, r2=0.006, p<0.05). In a post hoc analysis to explore the effects of diagnosis, in the broad psychosis group, the same direction of effect was observed (B=3.634, r2=0.012, p<0.01). This was also the case in the narrow psychosis group (SZ and SZA patients only) (B=3.578, r2=0.011, p<0.05). The association was in the same direction in the healthy participant only subgroup, but was nonsignificant. In patients, rerunning the analysis to include CPZ as a covariate did not affect the significance of the results observed. No association between verbal working memory, measured by LNS task, and rs2007044 was observed.
Table 2. Summary of Significant Findings from Regression Analyses in the Discovery Sample.
| rs ID | Test | Sample | n | Mean (SD) |
Bootstrap 95 CI |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| r2 | B | Lower | Upper | P | |||||||
| AA (n=251) | AG (n=330) | GG (n=133) | |||||||||
| CACNA1C | All | 714 | 35.17 (23.10) | 37.43 (25.55) | 41.61 (23.44) | 0.006 | 2.600 | 0.635 | 4.535 | 0.012* | |
| rs2007044 | SWM | Broad | 557 | 38.82 (19.93) | 41.91 (26.75) | 47.51 (21.30) | 0.012 | 3.634 | 1.133 | 6.184 | 0.006** |
| SZ/SZA | 409 | 40.72 (22.66) | 43.38 (25.76) | 49.94 (21.25) | 0.011 | 3.578 | 0.717 | 6.153 | 0.013* | ||
| Healthy | 157 | 15.73 (12.27) | 18.23 (16.42) | 21.27 (17.31) | 0.010 | 2.102 | −0.632 | 4.891 | 0.138 | ||
| GG (n=6) | GT (n=97) | TT (n=416) | |||||||||
| CACNB2 | All | 519 | 17.17 (1.72) | 16.43 (2.20) | 15.66 (3.25) | 0.012 | −0.772 | −1.210 | −0.337 | 0.003** | |
| rs7893279 | Hinting Task | Broad | 380 | 16.25 (0.96) | 16.00 (2.37) | 15.34 (3.55) | 0.006 | −0.643 | −1.203 | −0.122 | 0.021* |
| SZ/SZA | 295 | 16.33 (1.15) | 16.02 (2.38) | 15.15 (3.74) | 0.009 | −0.826 | −1.553 | −0.147 | 0.017* | ||
| Healthy | 139 | 19.00 (1.41) | 17.31 (1.51) | 16.59 (1.87) | 0.052 | −0.895 | −1.448 | −0.282 | 0.005** | ||
| TT (n=63) | TC (n=266) | CC (n=224) | |||||||||
| RIMS1 | All | 553 | 22.56 (6.36) | 22.32 (6.17) | 23.51 (5.83) | 0.008 | 0.831 | 0.300 | 1.568 | 0.024* | |
| rs1339227 | Eyes Test | Broad | 392 | 20.20 (6.12) | 20.51 (6.25) | 22.48 (6.14) | 0.018 | 1.272 | 0.235 | 2.228 | 0.007** |
| SZ/SZA | 273 | 18.84 (6.39) | 19.41 (6.25) | 22.28 (6.36) | 0.034 | 1.807 | 0.758 | 2.905 | 0.003** | ||
| Healthy | 161 | 28.00 (2.21) | 26.31 (3.62) | 26.37 (3.56) | 0.002 | −0.251 | −0.977 | 0.496 | 0.501 | ||
Abbreviations: SWM, CANTAB Spatial Working Memory (Errors); SZ/SZA, Schizophrenia/Schizoaffective Disorder; Broad, broad sense psychosis diagnosis of bipolar disorder with psychotic features, major depressive disorder with psychotic features, or psychosis not otherwise specified; includes SZ/SZA patients.
The r2 represents the percentage of variance for each neuropsychological test score explained by the genotype; B is regression coefficient; p-values uncorrected. Bold indicates significance at P<0.05, *P<0.05 and **P<0.01.
For the SNP rs7893279 (located within the gene encoding the calcium channel subunit B2 (CACNB2)) increased risk ‘T’ allele dosage was associated with lower accuracy on the Hinting Task, a measure of ToM ability (B=−0.772, r2=0.012, p<0.01). When the broad psychosis group was considered separately, this association remained significant (B=−0.643, r2=0.006, p<0.05). This association also remained significant in the narrow psychosis group (B=−0.826, r2=0.009, p<0.05) and in the healthy sample (B=−0.895, r2=0.052, p<0.01). No associations with other social cognition measures or measures of other cognitive domains were observed.
For rs1339227 (located ~50 kb from RIMS1, the only known gene at this locus) increased risk ‘C’ allele dosage was, contrary to our expectation, associated with improved Eyes Task performance, another measure of ToM ability (B=0.831, r2=0.01, p=0.02). When the broad psychosis group was considered separately, this association remained significant (B=1.272, r2=0.018, p<0.01). This association also remained significant in the narrow psychosis group with the amount of variance explained increasing (B=1.807, r2=0.034, p<0.01), suggesting that the genotype effects were more pronounced in this group. No significant association was observed in the healthy participants sample only.
Replication Analysis
We sought replication of our SWM and rs2007044 risk G genotype association (uncorrected). In the Cardiff replication sample (consisting of patients only, n=528), we sought to replicate this association between CACNA1C and working memory using the working memory domain score (calculated from both the Wechsler Memory Scale Spatial Span subtest and the LNS subtest) from the MATRICS battery. Based on this analysis (three genotype groups AA, AG, and GG), increased rs2007044 risk ‘G’ allele dosage was again associated with decreased working memory accuracy (B=−0.157, r2=0.01; p=0.03) Because the working memory domain score in the MATRICS is composed of verbal (LNS) and spatial (Spatial Span) components, we conducted a post hoc analysis to establish whether our result was being driven by either the verbal or spatial subtest. This analysis suggested that the association observed was driven by the verbal subtest performance (LNS: B=−0.200, 95% CI −0.358 to −0.042, p=0.013) as the spatial subtest by itself was nonsignificant (Spatial Span: B=−0.089, 95% CI −0.222 to 0.045, p=0.192).
In the German samples, two working memory measures were available. The first consisted of performance on an n-back test (1- and 2-back, a task that requires working memory to be constantly updated), and a composite working memory score based on the Wechsler Digit Span and Spatial Span Tasks (short-term maintenance tasks for working memory). Based on the same analysis undertaken as in the Irish and Cardiff sample, a main effect of CACNA1C rs2007044 genotype for the n-back test (2-back condition only) was observed such that the risk ‘G’ genotype carriers scored significantly below the nonrisk carriers in the patients (n=847; B=−2.615, r2=0.012; p=0.03) but not in healthy participants (n=342). No association between CACNA1C and the Digit/Spatial Span score was observed. For significant working memory associations in the replication samples, the effect size in both the Welsh (d=0.20) and German patient samples (d=0.22) was comparable to that observed in the Irish patient samples (d=0.22). Supplementary Table S5 provides the results of each cognitive test for this SNP to allow comparison of the specificity of the SWM association relative to other cognitive domains for rs2007044 (CACNA1C).
Finally, for the associations between social cognition and CACNB2 (Hinting task performance) and RIMS1 (Eyes task performance), neither risk variant was associated with social cognition as indexed by the MSCEIT ‘managing emotions’ subtest in the Cardiff samples, and no social cognition data were available in the German data set.
fMRI Spatial Working Memory Analysis
As we observed an effect of the rs2007044 risk ‘G’ genotype on working memory in two replication samples as well as our own, we also brought this SNP forward to investigate whether or not there was an association with altered connectivity during a SWM fMRI task. There were no significant differences between genotype groups for age, years of education, SWM accuracy, SWM reaction time, or gender in the MRI sample (p>0.05) (Table 3).
Table 3. CACNA1C Spatial Working Memory fMRI Study: Participant Demographics.
| Mean (SD) age | Mean (SD) years education | SWM mean (SD) accuracy | SWM mean (SD) reaction time | Gender (M/F) | |
|---|---|---|---|---|---|
| AA (N=25) | 30.88 (11.02) | 16.96 (3.84) | 64.80 (6.86) | 9070.61 (2332.96) | 15 : 10 |
| AG (N=42) | 27.86 (7.10) | 17.75 (3.24) | 62.17 (10.90) | 8535.44 (2063.93) | 15 : 27 |
| GG (N=17) | 29.00 (10.30) | 17.66 (2.87) | 64.94 (4.63) | 8811.90 (2296.80) | 7 : 10 |
| F or χ2 | F=0.868 | F=0.438 | F=0.972 | F=0.475 | χ2=3.822 |
| P | 0.424 | 0.647 | 0.383 | 0.624 | 0.148 |
Abbreviation: SWM, spatial working memory.
F-statistic: one-way ANOVA comparing age, years of education, spatial working memory accuracy, and reaction time between genotype groups; χ2 analysis: comparing gender by genotype group.
Neural Activity Associated with Spatial Working Memory Performance in Healthy Participants
Spatial working memory (1 dot and 3 dots combined) was associated with significantly increased neural activation in the frontoparietal attention network across our sample (Fox et al, 2005; Toro et al, 2008) including, bilaterally, the DLPFC (BA 9 and 46; tmax(84)=19.48; p<0.05 corrected; see Supplementary Appendix 1). In addition, the 3-dot condition was associated with increased activity across several of these frontal and parietal regions compared with the 1-dot condition (tmax(84)=12.99; p<0.05, corrected; see Supplementary Appendix 1). There were no significant differences between genotypes on either contrast, ie, genotypes did not influence BOLD response for the SWM task.
Functional Connectivity Analysis
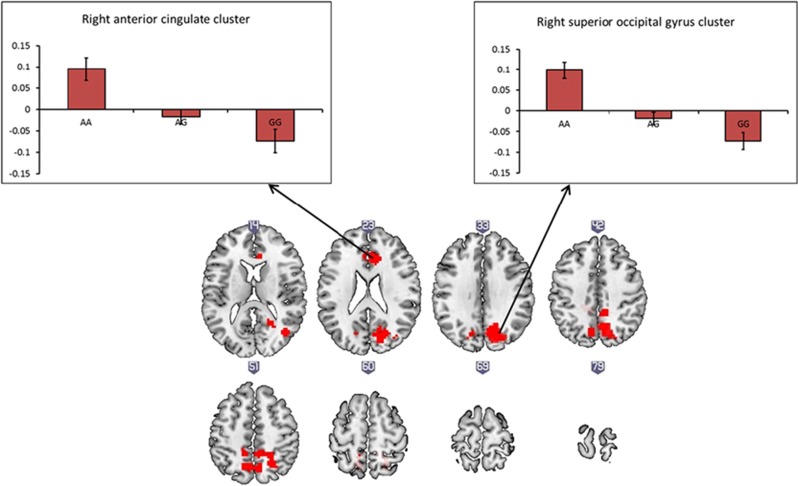
Decreased functional connectivity was associated with risk genotype, between the right DLPFC and three clusters incorporating the right cuneus, right anterior cingulate cortex, and left inferior frontal gyrus (tmax(82)=5.32; p<0.05, corrected; see Supplementary Appendix I1). As a QC measure, average parameter estimates were calculated for each individual, based on which five outliers were identified; after removing these outliers, however, findings were largely unchanged, with altered connectivity between the DLPFC and the right superior occipital gyrus/right cuneus and the right anterior cingulate cortex remaining significant (tmax(77) =4.92; p<0.05, corrected; see Table 4 and Figure 1). Finally, we examined functional connectivity of the right DLPFC across our sample using the ‘next local maximum’ approach (see Supplementary Appendix 1). Results of this analysis were again supportive of the main analysis, with altered connectivity between the rDLPFC and occipital and cingulate clusters observed, although at a lower threshold (p<0.001, uncorrected). To confirm at a group level that motion was not a confound in the genotype comparisons, we examined differences in motion across the three rs2007044 (CACNA1C) genotype groups. There were no significant differences in mean translation or mean rotation between the three genotype groups (see Supplementary Appendix 1 for further information).
Table 4. Clusters, Including Individual Peaks, Showing Significantly Decreased Functional Connectivity with the Right DLPFC with Increasing rs2007044 Risk Alleles: Global Maximum Approach, Five Outliers Removed (n=77).
| Cluster | Extent (voxels) | P-valuea | Cluster peaks | t-value | Z-value | Peak coordinates (MNI) |
|---|---|---|---|---|---|---|
| 1 | 888 | <0.001 | Right superior occipital gyrus | 4.92 | 4.55 | 21 −73 31 |
| Right cuneus | 4.65 | 4.33 | 15 −67 34 | |||
| Right precuneus | 4.45 | 4.17 | 9 −43 46 | |||
| 2 | 121 | 0.02 | Right anterior cingulate cortex | 4.90 | 4.53 | 6 29 19 |
The p-values are FWE-corrected for multiple comparisons at the cluster level.
Figure 1.
Effects of CACNA1C rs2007044 variant on right prefrontal connectivity during spatial working memory, global maximum approach, five outliers removed (n=77). The y-axis represents mean parameter estimates in arbitrary units; clusters are significant at p<0.05, FWE-corrected at the cluster level, and represent regions where functional connectivity with the right DLPFC is weaker with increasing risk G alleles. Each 2D axial slice is labeled with an x-coordinate (MNI space). Clusters are rendered on the ‘ch256’ brain template using MRIcroGL (http://www.mccauslandcenter.sc.edu/mricrogl/). Additional editing of figure (eg, changing the size/resolution) performed using MS Paint and/or Paint.NET v3.5.10.
Discussion
In this study we assessed the effects of 11 SZ-associated SNPs linked with synaptic transmission genes on behavioral and cortical measures of cognition. Three SNPs were found to be significantly associated with neuropsychological test scores, two of which are linked to calcium channel genes, and one with vesicular trafficking. Carriers of the SZ-associated risk ‘G’ allele at rs2007044 (located in intron 3 of CACNA1C) made more errors on a SWM task than nonrisk carriers in a risk allele dose-dependent manner, an effect seemingly driven by the patient sample. The same risk allele was also associated with poorer working memory performance in independent patient samples, although not for the same task as was associated with the Irish samples. No behavioral working memory associations were found with healthy participants in any sample. Finally, fMRI connectivity analysis of this risk allele in a subset of the discovery healthy participant sample suggested an allele dosage effect of the rs2007044 risk G allele on connectivity between the right DLPFC and multiple other cortical regions.
As reviewed by Heyes et al (2015) the role of voltage-gated calcium channels in increasing risk of psychiatric disorders including SZ is currently the focus of intense investigation. Multiple intronic variants situated within the gene coding for CACNA1C (eg, rs2007044, rs1006737, rs4765905) have been found to confer transdiagnostic susceptibility to SZ, bipolar disorder, and major depressive disorder (Cross-Disorder Group of the Psychiatric Genomics Consortium et al, 2013; Green et al, 2010; Nyegaard et al, 2010; The Schizophrenia Psychiatric GWAS Consortium, 2011; Williams et al, 2011; Zheng et al, 2014). Timothy Syndrome, a disorder that includes cognitive deficits (Splawski et al, 2004), has also been associated with a missense mutation in CACNA1C. A previously identified intronic CACNA1C risk variant rs1006737 (Nyegaard et al, 2010; Sklar et al, 2008), which is in moderately high LD with the PGC-identified variant rs2007044 investigated here (LD r2=0.79), was also previously associated with poorer cognitive performance in SZ patients and controls (Hori et al, 2012; Soeiro-de-Souza et al, 2013; Zhang et al, 2012). In the first study that reports on the cognitive effects of rs2007044, the results we observe across three independent cohorts are highly consistent with those reported for rs1006737 by Hori et al (2012) and Chen et al (2012), although one finding reported by Frazier et al (2014) reports the rs1006737 risk allele as being associated with an increase in cognitive function.
Neural Mechanism of CACNA1C Role in Working Memory
In seeking to establish the cortical basis of the CACNA1C rs2007044 risk allele effects on working memory, we analyzed fMRI data obtained during working memory task performance. Based on a previously reported methodology for assessing task-dependent functional connectivity (Paulus et al, 2013), we found that rs2007044 risk genotype was associated with decreased connectivity between the DLPFC and a distributed network of cortical regions (including the right cuneus, right anterior cingulate cortex, and left inferior frontal gyrus). This association suggested that during working memory task performance, risk carriers had difficulty with top-down initiated cortical responses that resulted from dysconnectivity between the right DLPFC and other frontal and parietal regions. Although no genotype effects were evident for neural activation (as measured by BOLD activations), it has previously been suggested that functional connectivity between brain regions may represent a more sensitive intermediate phenotype in identifying neural circuits for SZ risk variants compared with measures of neural activity (Meyer-Lindenberg, 2009). Previous connectivity analysis for rs1006737 in working memory also reported altered functional connectivity between the right DLPFC and the hippocampal formation; however, the results obtained by Paulus et al (2014), in which the rs1006737 risk allele (A) was associated with increased connectivity between the regions implicated, are inconsistent with the present study. Our results also differed from this previous study by suggesting that the pattern of altered connectivity between the right DLPFC and other cortical regions is more widespread. The reasons for this are uncertain and may relate to differences in either the SNP or working memory task studied. Whatever the cause, by again relating the deleterious effects of CACNA1C on working memory to prefrontal dysconnectivity, our study highlights calcium channel deregulation as an important mechanism that contributes to cognitive deficits. Differences in functional connectivity findings between studies are, as with other imaging analyses approaches, also likely to result from methodological differences (Bedenbender et al, 2011). As there is currently no gold standard method in functional connectivity research, these results should be interpreted with caution until they are replicated in an independent sample and shown to be consistent across different methodologies.
Molecular Mechanisms
CACNA1C codes for the α1c subunit of the L-type calcium channel that is particularly important to heart and brain function. In the brain, these channels are highly expressed in the dentate gyrus, a region synonymous with memory function (Jabès and Nelson, 2015; Kee et al, 2007). Although no significant findings in hippocampal regions are evident, the α1c channel subunit encoded by CACNA1C is also widely expressed throughout other areas of the brain, including regions of altered connectivity identified in the present and other studies. These α1c subunits are not only predominantly expressed postsynaptically on the cell soma but also on the spines and shafts of neuron dendrites (Zamponi et al, 2015), supporting neuronal plasticity, through which they are involved with learning and memory.
The molecular impact of rs2007044 has not yet been explored. For the rs1006737 SNP, risk genotype has been associated with increased CACNA1C mRNA expression in induced human neurons (Yoshimizu et al, 2015) and in human DLPFC (Bigos et al, 2010). In contrast, data from the Braineac database (Ramasamy et al, 2014) show that the SNP can have opposite effects—that CACNA1C expression is decreased in the presence of one or more risk (G) rs2007044 alleles—although this is dependent on brain region. It is currently unclear, therefore, how the behavioral and cortical SNP effects reported here might relate to difference in expression, and thereby to calcium subunit function, although this is likely to be influenced by the developmental stage and type of cell.
The two voltage-gated calcium channel genes included here play a key role in synaptic transmission. Calcium channels are composed of different channel-forming subunits. Each have multiple isoforms that determine specific cellular behavior (Breitenkamp et al, 2014). Contrary to the assumption that genes coding for calcium subunits might influence the same cognitive functions, the two calcium channel SNPs included here were found to influence different domains—working memory and ToM respectively. To our knowledge, no other group to date has examined the effects of this CACNB2 variant rs7893279 (or any other CACNB2 variant) on cognition, and it was not possible to replicate these findings in the independent samples available (see discussion of limitations below). At the same time, CACNA1C has recently also been associated with effects on social information processing (Dima et al, 2013). Therefore, whether this gene family has effects on more than one cognitive process warrants further investigation for potentially dissociable effects.
Study Limitations
Associations between common variants associated with risk for psychiatric disorders and certain phenotypes are expected to be small, and issues of both false positive and false negatives have been widely reported (Ioannidis et al, 2014). Here we sought to replicate our working memory finding in independent samples. Although a significant effect of rs2007044 on working memory was observed in the Irish, Welsh, and German samples, associations were with different working memory tasks in each sample. In the Irish patient samples, the association was with spatial but not verbal working memory. No association with the CPT was observed that, although included here as a measure of attention, is also significantly correlated with the CANTAB SWM and LNS working memory tasks. In the Cardiff sample (SZ/SZA patients only) the association observed was with a composite score, and in a follow-up analysis with verbal but not spatial working memory. Finally, in the German sample (SZ patients), the association was with n-back (2-back) performance but not with a second composite measure, or the individual tasks from which the composite score was derived. These between-sample differences between working memory tasks had not been hypothesized. It is possible that some measures may have differed in their sensitivity to gene effects in different cohorts. What is clear is that these differences cannot simply be attributed to verbal versus visuospatial effects as, eg, spatial effects were strongest in the Irish samples, but verbal effects were strongest in the Cardiff samples. The lack of association with other cognitive domains suggests that this association is just with working memory. At the same time, given that calcium channels have essential function in many types of neurons, and are expressed throughout the brain, and given previous studies of CACNA1C and cognition (eg, as reviewed by Rose and Donohoe, 2013) the cognitive effects of CACNA1C are unlikely to be cognitively or cortically specific.
The present study did not include patients in the fMRI analysis, thus requiring an inferential leap between the cognitive findings and the MRI findings. Confirming similar cortical effects in patients as found here in healthy participants will be important. Similarly, although the variant was associated with reduced working memory capacity in patients and healthy participants using neuropsychological assessment, no effects were observed in healthy participants on our fMRI SWM task. This is likely because of the reduced power in the MRI sample (n=77) to observe significant behavioral effects compared with the neuropsychological assessment sample. Nevertheless, this is an important limitation of the present study, limiting the extent to which we can infer that the variant affects working memory through a mechanism of altered prefrontal connectivity.
Although we have attempted to remove task-related variance out of the fMRI data using regression before connectivity analysis, it is important to note that task-related signal cannot be completely removed from BOLD data, making this an important limitation of the seed-voxel correlation approach to functional connectivity during task-based fMRI. Future studies could use PPI analysis to examine genetic effects on functional connectivity changes related to specific cognitive states (eg, SWM) (Friston et al, 1997). However, our current study, which is underpowered to detect smaller genetic effects, may not be sufficiently powered for PPI as this technique is associated with low statistical power and high incidence of false negatives (O'Reilly et al, 2012). In this study we sought to use methods as similar as possible to those of previous studies that have examined effects of CACNA1C variation on functional connectivity (eg, Bedenbender et al, 2011). However, it should be noted that the method of including peak voxel coordinates as nuisance covariates used here may be unsuccessful at fully removing effects of these differences in peak voxel coordinates if coordinates change in a quadratic rather than linear way. As such, future studies will be necessary to examine further the effects of differences in peak voxel coordinates on the estimation of functional connectivity.
Given the importance of social cognition to understanding functional disability in SZ (Green et al, 2015), our inability to replicate the association in our discovery sample between two other risk loci and ToM is noteworthy. In our discovery sample, based on a measure of ToM, we found that risk allele T carriers at rs7893279 scored lower on a ToM measure than noncarriers; and also that carriers of the C risk allele at rs1339227 scored higher on a ToM task than noncarriers, both for the group as a whole and for the SZ/SZA sample considered separately. The rs1339227 SNP is ~50 kb away from RIMS1; the closest gene in this region. The direction of this result was contrary to our expectation that risk allele dosage would be associated with poorer performance, and did not replicate in the Cardiff sample based on MSCEIT ‘managing emotions’ subtest performance; no social cognition measure was available in the German data set. The lack of either the same measure or any measure of social cognition in sufficiently large data sets to enable replication of genetic findings related to social cognition is currently an important limitation for our field, especially considering our surprising findings here.
Surprisingly, none of the SNPs identified as associated with cognitive performance in our discovery sample were linked to glutamate or dopaminergic transmission, given previous candidate gene studies (eg, COMT, DTNBP1). It is of interest to note that in a recent bioinformatics study seeking to prioritize signals identified by the Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014), Pers et al (2016) generated a gene set that overlapped substantially with the set reported here. This suggests that the findings observed through the novel approach taken here are unlikely to be accounted for purely on the basis of an idiosyncratic approach to gene prioritization. Finally, although this study was designed to test the relevance of genetic risk loci related to synaptic function as a potential biological mechanism of cognitive disability in SZ, this pathway in not unique in this regard. Other gene networks and biological pathways are also likely to be relevant, including gene sets with gene regulatory functions, eg, MIR137 (Cosgrove et al, 2017) and ZNF804A (Nicodemus et al, 2014) among others. Several other gene sets also warrant investigation, including gene sets with epigenetic effects and those with immune effects. In order to limit the SNPs included in the analysis, some synapse-related genes identified from the PGC-SZ were excluded if they were not involved in processes of neurotransmission, neurotransmitter secretion, or transduction as identified using the OMIM or Uniprot databases. As a more intensive investigation into each of the ~350 genes was not feasible, we took this broad approach to assigning function to genes from the PGC-SZ. A limitation of using this approach is that this may have led to the exclusion of some genes that may be expected, eg, from other databases, to be included under the function of synaptic transmission.
Conclusion
In conclusion, these data, based on previously identified SZ risk variants associated with synaptic transmission, provide some additional evidence for the role of CACNA1C in memory function already postulated in the literature. Furthermore, this association, we suggest, may result from patient risk carriers’ difficulty with top-down initiated responses caused by dysconnectivity between the right DLPFC and other cortical regions. These deficits, and the potential role of calcium channel subunits in contributing to these, are not effectively addressed by current therapeutics. In this context, the approach to characterizing the functional effects of risk variants at both a behavioral and neural systems level, reported here, may assist future efforts to identify novel biological targets for treatment.
Funding and disclosure
The authors declare no conflict of interest.
Acknowledgments
We thank all patients and their support staff and all healthy volunteers for participating in the data collection on which this manuscript is based. Recruitment and genotyping was supported by Science Foundation Ireland (SFI) (grants 12.IP.1359 and 08/IN.1/B1916) and the Wellcome Trust Case Control Consortium 2 project (grant 085475/B/08/Z) and the Wellcome Trust (grants 072894/Z/03/Z, 090532/Z/09/Z, and 075491/Z/04/B). The funding bodies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
The Wellcome Trust Case Control Consortium 2 Investigators
Peter Donnelly, Lesley Bates, Ines Barroso, Jenefer M. Blackwell, Elvira Bramon, Matthew A. Brown, Juan P. Casas, Aiden Corvin, Panos Deloukas, Audrey Duncanson, Janusz Jankowski, Hugh S. Markus, Christopher G. Mathew, Colin N. A. Palmer, Robert Plomin, Anna Rautanen, Stephen J. Sawcer, Richard C. Trembath, Ananth C. Viswanathan, Nicholas W. Wood, Chris C. A. Spencer, Gavin Band, Céline Bellenguez, Colin Freeman, Garrett Hellenthal, Eleni Giannoulatou, Lucinda Hopkins, Matti Pirinen, Richard Pearson, Amy Strange, Zhan Su, Damjan Vukcevic, Cordelia Langford, Sarah E. Hunt, Sarah Edkins, Rhian Gwilliam, Hannah Blackburn, Suzannah J. Bumpstead, Serge Dronov, Matthew Gillman, Emma Gray, Naomi Hammond, Alagurevathi Jayakumar, Owen T. McCann, Jennifer Liddle, Simon C. Potter, Radhi Ravindrarajah, Michelle Ricketts, Matthew Waller, Paul Weston, Sara Widaa, and Pamela Whittaker.
Supplementary Material
References
- Andreasen N (1995). Symptoms, signs, and diagnosis of schizophrenia. Lancet 346: 477–481. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S, Wheelwright S, Hill J, Raste Y, Plumb I (2001). The ‘Reading the Mind in the Eyes’ test revised version: a study with normal adults, and adults with Asperger syndrome or high-functioning autism. J Child Psychol Psychiatry 42: 241–251. [PubMed] [Google Scholar]
- Bedenbender J, Paulus FM, Krach S, Pyka M, Sommer J, Krug A et al (2011). Functional connectivity analyses in imaging genetics: considerations on methods and data interpretation. PLoS ONE 6: e26354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B et al (2010). Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry 67: 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenkamp A, Matthes J, Nass RD, Sinzig J, Lehmkuhl G, Nürnberg P et al (2014). Rare mutations of CACNB2 found in autism spectrum disease-affected families alter calcium channel function. PLoS ONE 9: e95579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott JH, Bertolino A, Mattay VS, Langheim FJ, Duyn J, Coppola R et al (2000). Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cereb Cortex 10: 1078–1092. [DOI] [PubMed] [Google Scholar]
- Cambridge Cognition (2016). CANTAB® [Cognitive assessment software]. All rights reserved www.cantab.com.
- Chen L, Wang X, Wang H, Li Y, Yan W, Han L et al (2012). miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer 48: 3104–3111. [DOI] [PubMed] [Google Scholar]
- Corcoran R, Mercer G, Frith CD (1995). Schizophrenia, symptomatology and social inference: investigating ‘theory of mind’ in people with schizophrenia. Schizophr Res 17: 5–13. [DOI] [PubMed] [Google Scholar]
- Cornblatt BA, Risch NJ, Faris G, Friedman D, Erlenmeyer-Kimling L (1988). The Continuous Performance Test, identical pairs version (CPT-IP): I. New findings about sustained attention in normal families. Psychiatry Res 26: 223–238. [DOI] [PubMed] [Google Scholar]
- Cosgrove D, Harold D, Mothersill O, Anney R, Hill MJ, Bray NJ et al (2017). MiR-137-derived polygenic risk: effects on cognitive performance in patients with schizophrenia and controls. Transl Psychiatry 7: e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics ConsortiumCross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Cross-Disorder Group of the Psychiatric Genomics Consortium, Ripke S, Cross-Disorder Group of the Psychiatric Genomics Consortium, Neale BM, Cross-Disorder Group of the Psychiatric Genomics Consortium, Faraone SV, Cross-Disorder Group of the Psychiatric Genomics Consortium, Purcell SM et al (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima D, Jogia J, Collier D, Vassos E, Burdick KE, Frangou S (2013). Independent modulation of engagement and connectivity of the facial network during affect processing by CACNA1C and ANK3 risk genes for bipolar disorder. JAMA Psychiatry 70: 1303–1311. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE et al (2001). Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 98: 6917–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First M, Spitzer R, Gibbon M, Williams J (2002) Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition (SCID-I/P). New York State Psychiatric Institute: New York, NY. [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME (2005). The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA 102: 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TW, Youngstrom EA, Frankel BA, Zunta-Soares GB, Sanches M, Escamilla M et al (2014). Candidate gene associations with mood disorder, cognitive vulnerability, and fronto-limbic volumes. Brain Behav 4: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Buechel C, Fink G, Morris J, Rolls E, Dolan R (1997). Psychophysiological and modulatory interactions in neuroimaging. Neuroimage 6: 218–229. [DOI] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S et al (2010). The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry 15: 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MF, Horan WP, Lee J (2015). Social cognition in schizophrenia. Nat Rev Neurosci 16: 620–631. [DOI] [PubMed] [Google Scholar]
- Hargreaves A, Anney R, O'Dushlaine C, Nicodemus KK et alSchizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ), Wellcome Trust Case Control Consortium 2 (2014). The one and the many: effects of the cell adhesion molecule pathway on neuropsychological function in psychosis. Psychol Med 44: 2177–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härting C, Markowitsch HJ, Neufeld H, Calabrese P, Deisinger K, Kessler J (2000) Wechsler Gedächtnis Test-Revidierte Fassung (WMS-R). Huber: Bern. [Google Scholar]
- Heyes S, Pratt WS, Rees E, Dahimene S, Ferron L, Owen MJ et al (2015). Genetic disruption of voltage-gated calcium channels in psychiatric and neurological disorders. Prog Neurobiol 134: 36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H, Yamamoto N, Fujii T, Teraishi T, Sasayama D, Matsuo J et al (2012). Effects of the CACNA1C risk allele on neurocognition in patients with schizophrenia and healthy individuals. Sci Rep 2: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM (2012) IBM SPSS Statistics for Windows version 21.0 IBM: Armonk, NY. [Google Scholar]
- Ioannidis JP, Munafo MR, Fusar-Poli P, Nosek BA, David SP (2014). Publication and other reporting biases in cognitive sciences: detection, prevalence, and prevention. Trends Cogn Sci 18: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabès A, Nelson CA (2015). 20 years after ‘The ontogeny of human memory A cognitive neuroscience perspective,’ where are we? Int J Behav Dev 39: 293–303. [Google Scholar]
- Kee N, Teixeira CM, Wang AH, Frankland PW (2007). Preferential incorporation of adult-generated granule cells into spatial memory networks in the dentate gyrus. Nat Neurosci 10: 355–362. [DOI] [PubMed] [Google Scholar]
- Kinderman P, Bentall RP (1996). A new measure of causal locus: the internal, personal and situational attributions questionnaire. Pers Individ Dif 20: 261–264. [Google Scholar]
- Meyer-Lindenberg AS (2009). Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp 30: 1938–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus K, Hargreaves A, Morris D, Anney R, Gill M, Corvin A et al (2014). Variability in working memory performance explained by epistasis vs polygenic scores in the ZNF804A pathway. JAMA Psychiatry 71: 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuechterlein KH, Asarnow R (2004) 3-7 Continuous Performance Test. University of California, Los Angeles: Los Angeles, CA. [Google Scholar]
- Nuechterlein KH, Green MF (2006) MATRICS Consensus Cognitive Battery. Manual MATRICS Assessment: Los Angeles, CA. [Google Scholar]
- Nyegaard M, Demontis D, Foldager L, Hedemand A, Flint TJ, Sorensen KM et al (2010). CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry 15: 119–121. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V et al (2008). Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 40: 1053–1055. [DOI] [PubMed] [Google Scholar]
- O'Reilly JX, Woolrich MW, Behrens TE, Smith SM, Johansen-Berg H (2012). Tools of the trade: psychophysiological interactions and functional connectivity. Soc Cogn Affect Neurosci 7: 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMIM (2014) McKusick-Nathans Institute of Genetic Medicine. Johns Hopkins University: Baltimore, MD. [Google Scholar]
- Park J-H, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ et al (2010). Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet 42: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus FM, Bedenbender J, Krach S, Pyka M, Krug A, Sommer J et al (2014). Association of rs1006737 in CACNA1C with alterations in prefrontal activation and fronto-hippocampal connectivity. Hum Brain Mapp 35: 1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus FM, Krach S, Bedenbender J, Pyka M, Sommer J, Krug A et al (2013). Partial support for ZNF804A genotype-dependent alterations in prefrontal connectivity. Hum Brain Mapp 34: 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pers TH, Timshel P, Ripke S, Lent S, Sullivan PF, O'Donovan MC et al (2016). Comprehensive analysis of schizophrenia-associated loci highlights ion channel pathways and biologically plausible candidate causal genes. Hum Mol Genet 25: 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocklington AJ, O'Donovan M, Owen MJ (2014). The synapse in schizophrenia. Eur J Neurosci 39: 1059–1067. [DOI] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al (2014). Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17: 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson IH, Manly T, Andrade J, Baddeley BT, Yiend J (1997). Oops!': performance correlates of everyday attentional failures in traumatic brain injured and normal subjects. Neuropsychologia 35: 747–758. [DOI] [PubMed] [Google Scholar]
- Rose EJ, Donohoe G (2013). Brain vs behavior: an effect size comparison of neuroimaging and cognitive studies of genetic risk for schizophrenia. Schizophr Bull 39: 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose EJ, Greene C, Kelly S, Morris DW, Robertson IH, Fahey C et al (2012. a). The NOS1 variant rs6490121 is associated with variation in prefrontal function and grey matter density in healthy individuals. Neuroimage 60: 614–622. [DOI] [PubMed] [Google Scholar]
- Rose EJ, Morris DW, Fahey C, Robertson IH, Greene C, O'Doherty J et al (2012. b). The effect of the neurogranin schizophrenia risk variant rs12807809 on brain structure and function. Twin Res Hum Genet 15: 296–303. [DOI] [PubMed] [Google Scholar]
- Rose EJ, Morris DW, Hargreaves A, Fahey C, Greene C, Garavan H et al (2013). Neural effects of the CSMD1 genome-wide associated schizophrenia risk variant rs10503253. Am J Med Genet B Neuropsychiatr Genet 162B: 530–537. [DOI] [PubMed] [Google Scholar]
- Sarter M, Bruno JP, Parikh V (2007). Abnormal neurotransmitter release underlying behavioral and cognitive disorders: toward concepts of dynamic and function-specific dysregulation. Neuropsychopharmacology 32: 1452–1461. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K et al (2008). Whole-genome association study of bipolar disorder. Mol Psychiatry 13: 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeiro-de-Souza M, Bio D, Dias V, Vieta E, Machado-Vieira R, Moreno R (2013). The CACNA1C risk allele selectively impacts on executive function in bipolar type I disorder. Acta Psychiatr Scand 128: 362–369. [DOI] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R et al (2004). Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell 119: 19–31. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2015). Gene Ontology Consortium: going forward. Nucleic Acids Res 43(D1): D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Schizophrenia Psychiatric GWAS Consortium (2011). Genome-wide association study identifies five new schizophrenia loci. Nat Genet 43: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Fox PT, Paus T (2008). Functional coactivation map of the human brain. Cereb Cortex 18: 2553–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2014). UniProt: a hub for protein information. Nucleic Acids Res 43(Database issue): D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JT, Corvin A, Owen MJ, Williams H, Dragovic M, Quinn EM et al (2010). Psychosis susceptibility gene ZNF804A and cognitive performance in schizophrenia. Arch Gen Psychiatry 67: 692–700. [DOI] [PubMed] [Google Scholar]
- Wechsler D (1997. a) WAIS-III, Wechsler Adult Intelligence Scale: Administration and Scoring Manual. Psychological Corporation: London. [Google Scholar]
- Wechsler D (1997. b) Wechsler Memory Scale (WMS-III). Psychological Corporation: London. [Google Scholar]
- Wechsler TUH-RH (1991) Intelligenztest für Erwachsene Revision. Göttingen: Hogrefe. [Google Scholar]
- Williams HJ, Craddock N, Russo G, Hamshere ML, Moskvina V, Dwyer S et al (2011). Most genome-wide significant susceptibility loci for schizophrenia and bipolar disorder reported to date cross-traditional diagnostic boundaries. Hum Mol Genet 20: 387–391. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T, Pan JQ, Mungenast AE, Madison JM, Su S, Ketterman J et al (2015). Functional implications of a psychiatric risk variant within CACNA1C in induced human neurons. Mol Psychiatry 20: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamponi GW, Striessnig J, Koschak A, Dolphin AC (2015). The physiology, pathology, and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharmacol Rev 67: 821–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shen Q, Xu Z, Chen M, Cheng L, Zhai J et al (2012). The effects of CACNA1C gene polymorphism on spatial working memory in both healthy controls and patients with schizophrenia or bipolar disorder. Neuropsychopharmacology 37: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F, Zhang Y, Xie W, Li W, Jin C, Mi W et al (2014). Further evidence for genetic association of CACNA1C and schizophrenia: new risk loci in a Han Chinese population and a meta-analysis. Schizophr Res 152: 105–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.