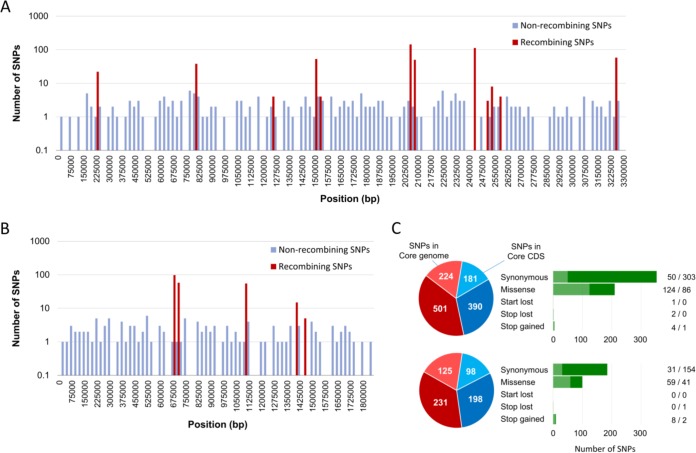
FIG 1 .
Frequency and distribution of SNPs in the core genome and core genes in the two chromosomes of the 48 V. parahaemolyticus strains used in this study. SNPs that were nonrecombinogenic and aroused by recombination are blue and red, respectively. (A) SNPs in chromosome I. (B) SNPs in chromosome II. (C) Summary of the frequencies of the SNPs found in each chromosome and the changes that occurred in the predicted amino acid sequence at that position (synonymous versus nonsynonymous, start or stop codons). Chromosome I, top; chromosome II, bottom. Light red, recombining SNPs; dark red, nonrecombining SNPs in core genome; light blue, recombining SNPs; dark blue, nonrecombining SNPs in CDS regions.