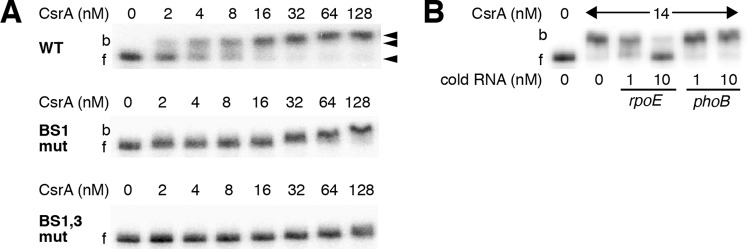
FIG 3.
Gel mobility shift analysis of CsrA-rpoE leader RNA interaction. (A) Labeled rpoE RNA (40 pM) was incubated with the concentration of CsrA shown at the top of each lane. Positions of bound (b) and free (f) RNA are marked with arrowheads. Experiments were performed at least twice, and representative gels are shown. CsrA formed a high-affinity complex with WT rpoE RNA and a second complex at higher protein concentrations. CsrA binding was greatly reduced when the GGA motif of binding site 1 (BS1 mut) was changed to CCA, and it was eliminated when combined with a GGA-to-CCA mutation in BS3 (BS1,3 mut). (B) RNA competition experiment demonstrating binding specificity. Labeled rpoE RNA (0.1 nM) was combined with the indicated concentration of unlabeled specific (rpoE) or nonspecific (phoB) competitor RNA and incubated with the concentration of CsrA shown at the top of each lane. Positions of bound (b) and free (f) RNA are marked. Experiments were performed twice, and a representative gel is shown.