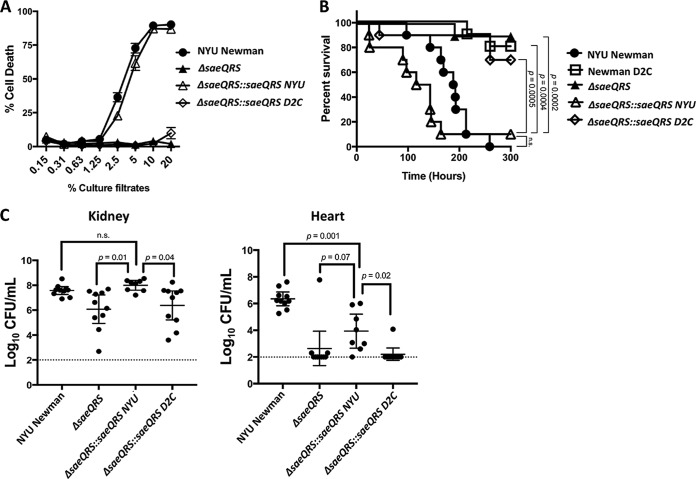
FIG 5.
SNP C595T in Newman D2C abolishes pathogenesis. (A) The NYU Newman saeR allele restored cytotoxicity to an ΔsaeQRS mutant, whereas the saeQRS allele from Newman D2C provided no restoration of cytotoxicity (n = 6 donors). Intoxications were repeated with unique transductants for both ΔsaeQRS::saeQRS D2C and ΔsaeQRS::saeQRS NYU, and the same results were achieved (data not shown). (B) Mice infected intravenously were susceptible to acute lethality only in the presence of the NYU Newman saeQRS allele, as neither Newman D2C, ΔsaeQRS, nor ΔsaeQRS::saeQRS D2C had acutely lethal effects on the mice (n = 10 mice/strain). The points indicate individual mouse deaths. Infections were done in sets of 5 mice, and each infection was performed with a unique transductant for ΔsaeQRS::saeQRS D2C and ΔsaeQRS::saeQRS NYU. Statistical analysis for survival was performed using the Gehan-Breslow-Wilcoxon test with P values adjusted for multiple comparisons. (C) Mice infected intravenously exhibited reduced bacterial burdens in critical organs when infected with either ΔsaeQRS or ΔsaeQRS::saeQRS D2C compared to that of NYU Newman or ΔsaeQRS::saeQRS NYU (n = 10 mice/strain). Dotted lines represent the limit of detection for bacterial burden in these organs. Statistical analysis was performed using one-way analysis of variance (ANOVA) with post hoc Holm-Sidak multiple-comparison tests where appropriate. The error bars indicate standard errors of the means.