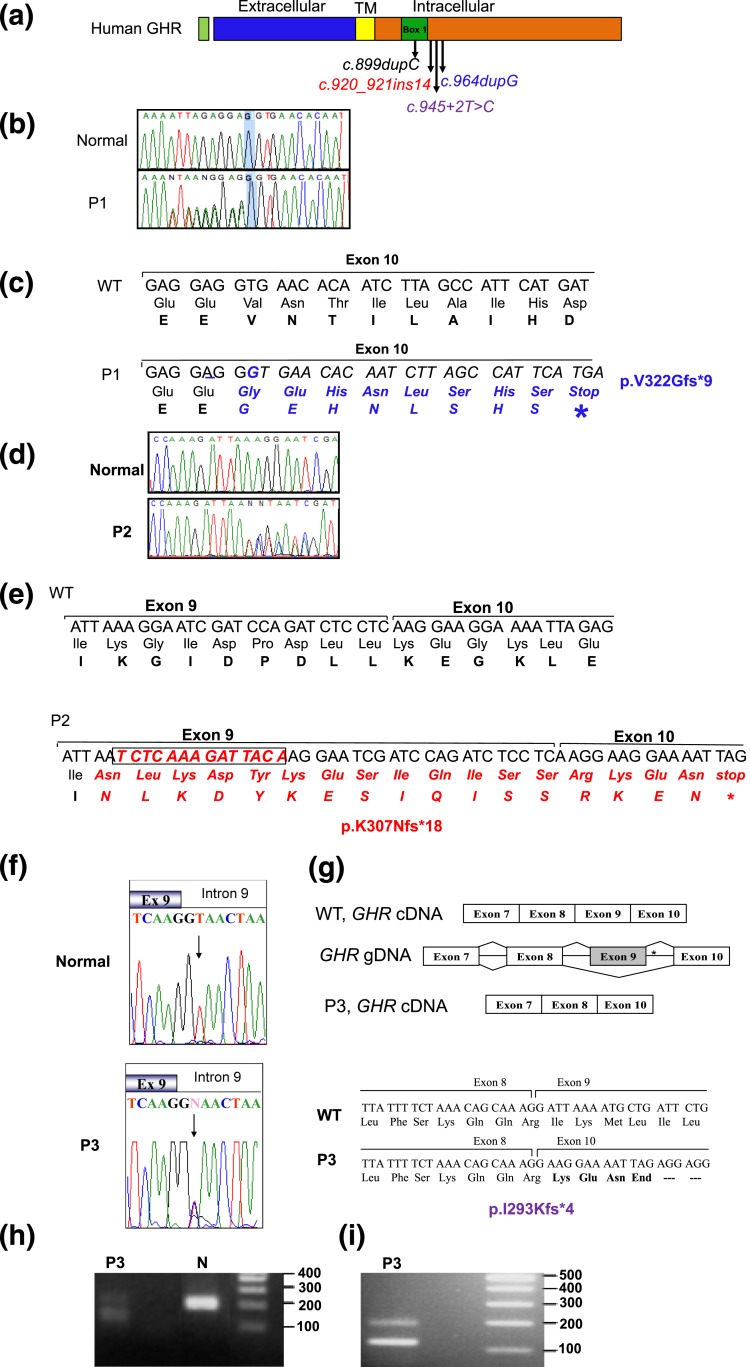
Figure 3.
Heterozygous GHR variants identified in affected patients. (a) Schematic representation of the hGHR protein showing the various domains and the location of the heterozygous mutations, c.899dupC, c.920_921ins14, c.945+2T>C, and c.964dupG. TM, transmembrane. (b) Electropherograms of the WT GHR and P1 carrying the c.964dupG variant. The reverse primer was used for sequencing and the electropherogram presented is the complement, forward sequences. Heterozygosity is, therefore, visualized 5′ to the highlighted G nucleotide. (c) DNA sequence (5′ to 3′) and corresponding amino acids showing the duplication of the nucleotide G (underlined) in the c.964dupG variant and the resulting frameshift and early termination (stop). (d) Electropherograms of the forward WT GHR and P2 showing the c.920_921ins14 variant. (e) DNA sequence (5′ to 3′) and corresponding amino acid sequence showing the insertion of 14 bp at nucleotide 921 of GHR cDNA (boxed) in the c.920_921ins14 variant, the resulting frameshift, and stop. (f) Electropherograms of the exon 9–intron 9 junction from WT GHR and P3 carrying the c.945+2T>C variant (arrow); the mother, but not the father, carried the variant, as did the maternal grandfather (data not shown). (g) Schematic, c.945+2T>C splicing out of exon 9. *Position of the mutation. The cDNA sequence and translated products of WT (GHR) and the c.945+2T>C variant are shown (bottom) along with the splice junctions. (h) DNA gel of GHR mRNA RT-PCR products from a normal control and P3, amplified using primers from exon 8 to 10. The expected product size is 187 bp (control). The presence of a smaller 117-bp band in the proband confirms deletion of exon 9. (i) A larger sample (20 µL) of the RT-PCR product better demonstrates the presence of two products (187 bp and 117 bp) carried by P3.