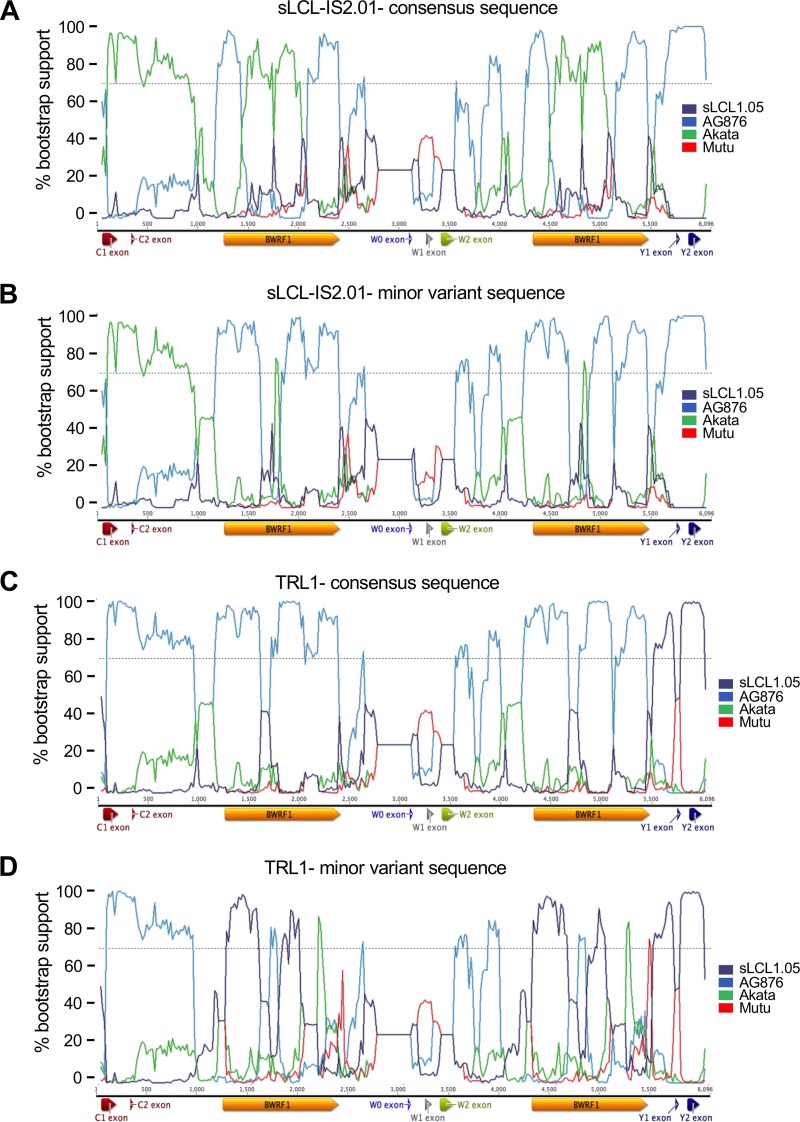
FIG 8.
Bootstrap analysis of genetic exchange in IR1 regions of sLCL-IS2.01 and TRL1. The consensus sequences for sLCL-IS2.01 (A) and TRL1 (C) were modified to contain all of the minor variant bases (B and D). These two sequences were compared to those of a set of four viruses from different clades on the phylogenetic tree to identify regions of similarity, using Bootscan. This analysis simulates phylogenetic tree assembly for a 200-bp sliding window, with the y axis indicating the percentage of occasions that each strain is the nearest neighbor on the tree and the x axis marking position, as illustrated by the IR1 schematic below each plot.