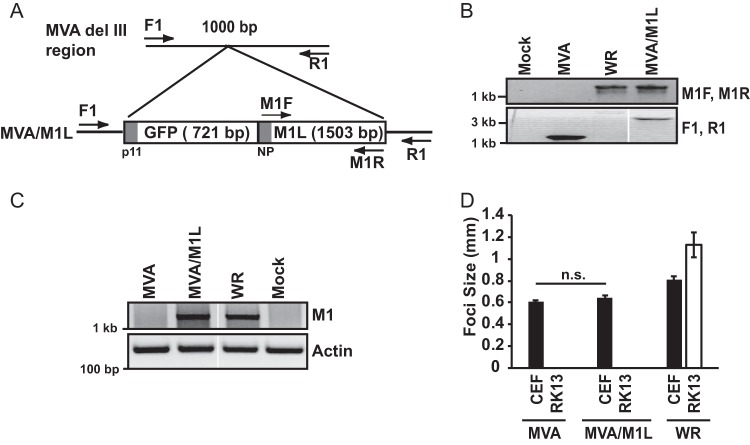
FIG 1.
Creation and characterization of an M1L-expressing MVA virus. (A) Schematic showing insertion of tandem GFP and M1L genes into the del III region of the MVA genome, in which the GFP gene is under the control of the poxvirus p11 promoter (p11) and the M1L gene is under the control of its natural promoter (NP). Primers used for verifying insertion of this GFP-M1L cassette are shown. (B) CEF monolayers were mock infected or infected with MVA, MVA/M1L, or WR (MOI = 10). At 24 h p.i., infected cells were harvested and lysed. DNA was subjected to PCR amplification using either the M1F and M1R primer set to amplify the M1L gene or the F1 and R1 primers to PCR amplify the MVA del III region. A portion of each PCR amplification reaction mixture was separated by agarose gel electrophoresis, and DNA was visualized using ethidium bromide staining. Reactions were analyzed in the same gel, and gel images were spliced for labeling purposes. (C) Detection of M1L gene transcription using semiquantitative RT-PCR. PMA-stimulated THP-1 cells were mock infected or infected with the indicated viruses (MOI = 2). At 6 h p.i., cells were collected and total RNA was extracted from lysed cells. Total RNA was reverse transcribed into cDNA. A portion of cDNA was incubated with primers either nested inside the M1L gene or for the actin gene as a control. A portion of each PCR was analyzed by agarose gel electrophoresis, and PCR amplicons were detected by using ethidium bromide staining of the gel. Reactions were analyzed in the same gel, and gel images were spliced for labeling purposes. (D) CEF or RK13 cellular monolayers were infected with MVA, MVA/M1L, or WR (50 PFU/well of a six-well plate). At 24 h p.i., cells were fixed and incubated in a solution containing anti-vaccinia virus antiserum, followed by a solution containing HRP-conjugated goat anti-rabbit antiserum. The diameters of at least 10 foci per condition were measured, and results are presented as the mean focus size ± SEM for each sample.