Table 2.
| A Summary of the total number of Bp enriched membrane fraction (EMF) proteins commonly and uniquely identified among the Bp strains assessed in the study. | |
|---|---|
| Strains | Total number of proteins |
|
| |
| T | 182 |
| C | 175 |
| D | 175 |
| F | 176 |
| Strain combinations | Total number of common proteins |
|
| |
| T, C, D, and F | 163 |
| T and C only | 1 |
| T and D only | 3 |
| T and F only | 0 |
| C and D only | 0 |
| C and F only | 0 |
| D and F only | 0 |
| T, C and D only | 1 |
| T, C and F only | 5 |
| T, D and F only | 2 |
| C, D and F only | 3 |
| Strains | Total number of unique proteins |
|
| |
| T only | 7 |
| C only | 2 |
| D only | 3 |
| F only | 3 |
| D Summary of Bp EMF proteins identified among Bp strains assessed in the study (** — Table 2A). | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Protein | Accession # |
ID | Gene | Size (kDa) |
Location | T | C | D | F | Function |
| 30S ribosomal protein S2 | NP_880161 | BP1419 | rpsB | 28 | C | 28 (4) | 23 (3) | 21 (3) | 23 (3) | Translation |
| 30S ribosomal protein S3 | NP_882129 | BP3619 | rpsC | 29 | C | 27 (4) | 27 (4) | 27 (5) | 27 (4) | Translation |
| 50S ribosomal protein L16 | NP_882130 | BP3620 | rplP | 15 | C | 26 (3) | 36 (4) | 19 (2) | 29 (3) | Translation |
| 50S ribosomal protein L18 | NP_882141 | BP3632 | rplR | 13 | C | 20 (2) | 20 (2) | 30 (3) | 20 (2) | Translation |
| 50S ribosomal protein L2 | NP_882126 | BP3616 | rplB | 30 | C | 14 (3) | 11 (2) | 11 (2) | 17 (3) | Translation |
| 50S ribosomal protein L5 | NP_882137 | BP3628 | rplE | 20 | C | 17 (3) | 12 (2) | 12 (2) | 18 (3) | Translation |
| 50S ribosomal protein L6 | NP_882140 | BP3631 | rplF | 19 | C | 27 (3) | 27 (3) | 27 (3) | 19 (2) | Translation |
| 2-Isopropylmalate synthase | NP_879030 | BP0131 | leuA | 62 | U | 6 (2) | 14 (5) | 8 (3) | 11 (4) | Metabolism |
| 5-Meta^ | NP_881170 | BP2543 | metA | 84 | U | 1 (1) | 4 (2) | 4 (2) | 4 (2) | Metabolism |
| ABC transporter | NP_879529 | BP0697 | U | 29 | CM | 22 (3) | 22 (3) | 27 (4) | 28 (4) | Membrane transport |
| Acteyl-CoA carboxylase carboxyltransferase subunit alpha | NP_880596 | BP1910 | accA | 35 | C | 17 (4) | 29 (6) | 24 (5) | 17 (4) | Metabolism |
| Aconitate hydratase | NP_880684 | BP2014 | acnA | 99 | C | 5 (3) | 5 (3) | 3 (2) | 4 (2) | Metabolism |
| Acriflavine resistance protein B | NP_879779 | BP0985 | acrB | 116 | CM | 6 (4) | 3 (2) | 5 (3) | 3 (2) | Drug resistance |
| Adenylosuccinate lyase | NP_881474 | BP2890 | purB | 50 | C | 7 (3) | 5 (2) | 9 (3) | 9 (3) | Metabolism |
| Alanyl-tRNA synthetase | NP_880538 | BP1836 | alaS | 96 | C | 15 (10) | 9 (6) | 8 (6) | 8 (5) | Translation |
| ATP-dependent protease La | NP_880488 | BP1777 | Ion | 90 | C | 5 (4) | 5 (3) | 6 (4) | 5 (3) | Metabolism |
| Autotransporter | NP_880953 | BP2315 | vag8 | 101 | OM/E | 24 (15) | 37 (22) | 28 (18) | 38 (22) | Transport |
| Autotransporter subtilisin-like protease | CAC44081 | U | sphB1 | 114 | OM/E | 14 (9) | 16 (9) | 23 (15) | 17 (10) | Transport |
| Bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | NP_880691 | BP2021 | acnB | 95 | C | 8 (5) | 6 (3) | 9 (6) | 6 (3) | Metabolism |
| Bifunctional hemolysin-adenylate cyclase precursor | NP_879578 | BP0760 | cyaA | 188 | E | 8 (8) | 18 (20) | 10 (12) | 14 (15) | Metabolism |
| Capsular polysaccharide biosynthesis protein | NP_880357 | BP1629 | wbpO | 47 | C | 13 (3) | 13 (3) | 10 (2) | 10 (2) | Metabolism |
| Cell division protein | NP_879861 | BP1077 | ftsH | 69 | CM | 20 (9) | 15 (6) | 12 (6) | 15 (7) | Cell division |
| Cell division protein FtsA | NP_881594 | BP3019 | ftsA | 45 | C | 18 (5) | 18 (5) | 17 (5) | 14 (4) | Cell division |
| Chain A, structure of the membrane protein Fhac | 2QDZ_A | U | U | 61 | OM | 22 (6) | 26 (8) | 25 (8) | 27 (9) | Transport |
| Chaperonin GroEL | NP_882014 | BP3495 | groEL | 60 | C | 16 (5) | 13 (5) | 20 (8) | 12 (5) | RNA degradation |
| Competence lipoprotein | NP_879922 | BP1146 | comL | 29 | OM | 21 (4) | 12 (2) | 26 (5) | 16 (3) | Transport |
| Cytochrome C1 precursor | NP_879155 | BP0275 | petC | 31 | U | 34 (6) | 24 (4) | 13 (3) | 29 (4) | Metabolism |
| d-fructose-6-phosphate amidotransferase^ | NP_879503 | BP0666 | glmS | 67 | C | 7 (3) | 4 (1) | 10 (4) | 8 (3) | Metabolism |
| Dihydrolipoamide acetyltransferase^ | NP_879904 | BP1125 | odhB | 44 | CM | 3 (1) | 24 (4) | 19 (3) | 11 (2) | Metabolism |
| Dihydroorotate dehydrogenase 2 | NP_881968 | BP3442 | pyrD | 38 | CM | 21 (4) | 18 (3) | 18 (3) | 13 (2) | Metabolism |
| Dihydroxy-acid dehydratase | NP_879169 | BP0289 | ilvD | 68 | C | 7 (3) | 5 (2) | 5 (2) | 5 (2) | Metabolism |
| DNA gyrase subunit B | NP_879342 | BP0489 | gyrB | 90 | C | 4 (2) | 7 (4) | 3 (2) | 5 (3) | DNA synthesis |
| DNA topoisomerase III^ | NP_879317 | BP0460 | topB | 96 | C | 9 (5) | 9 (5) | 8 (4) | 2 (1) | DNA synthesis |
| DNA-directed RNA polymerase subunit beta | NP_878932 | BP0015 | rpoB | 151 | C | 3 (2) | 7 (7) | 5 (5) | 4 (4) | Metabolism |
| DNA-directed RNA polymerase subunit beta | NP_878933 | BP0016 | rpoC | 155 | C | 3 (3) | 4 (4) | 3 (4) | 2 (2) | Metabolism |
| Elongation factor G | NP_882120 | BP3610 | fusA | 77 | C | 26 (11) | 24 (10) | 13 (6) | 20 (9) | Translation |
| Elongation factor Tu | NP_878925 | BP0007 | tuf | 44 | C | 38 (9) | 31 (7) | 21 (5) | 33 (8) | Translation |
| Enoyl- (acyl carrier protein) reductase^ | NP_881766 | BP3215 | fabL | 29 | CM | 5 (1) | 9 (2) | 9 (2) | 4 (1) | Lipid metabolism |
| F0F1 ATP synthase subunit alpha^ | NP_881828 | BP3286 | atpA | 56 | C | 8 (3) | 5 (2) | 11 (4) | 2 (1) | Energy metabolism |
| F0F1 ATP synthase subunit B | NP_881826 | BP3284 | atpF | 17 | CM | 30 (4) | 30 (4) | 43 (6) | 37 (5) | Energy metabolism |
| F0F1 ATP synthase subunit beta | NP_881830 | BP3288 | atpD | 51 | CM | 17 (5) | 23 (6) | 7 (2) | 15 (3) | Energy metabolism |
| Filamentous hemagglutinin/adhesion | NP_880571 | BP1879 | fhaB | 394 | OM | 18 (43) | 16 (37) | 16 (38) | 15 (34) | Adherence |
| Glutamate synthase [NADPH] large chain precursor | NP_882256 | BP3753 | qltb | 174 | C | 7 (7) | 3 (4) | 2 (2) | 6 (6) | Amino acid metabolism |
| Guanosine-3',5'-bis (diphosphate) 3'-pyrophosphohydrolase | NP_880309 | BP1576 | spoT | 83 | U | 5 (3) | 7 (4) | 5 (3) | 10 (6) | Nucleotide metabolism |
| Histone protein | NP_881561 | BP2985 | bpH1 | 19 | C | 18 (2) | 18 (2) | 18 (2) | 18 (2) | DNA synthesis |
| HlyD family secretion protein^ | NP_882313 | BP3815 | hlyD | 46 | CM | 10 (2) | 5 (1) | 13 (3) | 10 (2) | Signal transduction |
| Homoserine dehydrogenase^ | NP_881384 | BP2784 | U | 48 | C | 15 (4) | 18 (5) | 5 (1) | 18 (5) | Amino acid metabolism |
| Hypothetical protein BP0162 | NP_879055 | BP0162 | HP | 37 | U | 14 (3) | 9 (2) | 23 (4) | 23 (4) | HP |
| Hypothetical protein BP0205 | NP_879093 | BP0205 | HP | 21 | U | 44 (7) | 47 (8) | 39 (6) | 44 (7) | HP/transport |
| Hypothetical protein BP0325^ | NP_879200 | BP0325 | HP | 43 | CM | 3 (1) | 8 (3) | 3 (1) | 5 (2) | HP/membrane transport |
| Hypothetical protein BP0387 | NP_879258 | BP0387 | HP | 27 | P | 27 (4) | 21 (3) | 32 (5) | 30 (4) | HP |
| Hypothetical protein BP0606 | NP_879449 | BP0606 | HP | 15 | U | 23 (2) | 31 (3) | 31 (3) | 23 (2) | HP |
| Hypothetical protein BP1057 | NP_879842 | BP1057 | HP | 12 | U | 51 (3) | 23 (2) | 23 (2) | 39 (2) | HP |
| Hypothetical protein BP1426 | NP_880168 | BP1426 | HP | 49 | CM | 20 (5) | 11 (3) | 7 (2) | 11 (3) | HP |
| Hypothetical protein BP1438 | NP_880180 | BP1438 | HP | 16 | U | 16 (2) | 16 (2) | 15 (2) | 16 (2) | HP |
| Hypothetical protein BP1440 | NP_880182 | BP1440 | HP | 34 | U | 42 (8) | 36 (7) | 44 (8) | 42 (9) | HP/protein stability |
| Hypothetical protein BP1485 | NP_880222 | BP1485 | HP | 58 | C | 42 (12) | 25 (8) | 31 (9) | 31 (D) | HP/protein transport |
| Hypothetical protein BP1903 | NP_880589 | BP1903 | HP | 58 | CM | 5 (2) | 7 (2) | 7 (3) | 7 (2) | HP |
| Hypothetical protein BP2141^ | NP_880795 | BP2141 | HP | 17 | U | 9 (1) | 24 (2) | 24 (2) | 31 (3) | HP |
| Hypothetical protein BP2191 | NP_880839 | BP2191 | hflK | 48 | U | 35 (8) | 22 (5) | 32 (8) | 35 (9) | HP |
| Hypothetical protein BP2197 | NP_880845 | BP2197 | HP | 23 | U | 25 (3) | 47 (6) | 32 (4) | 35 (4) | HP |
| Hypothetical protein BP2323 | NP_880961 | BP2323 | HP | 28 | U | 24 (3) | 18 (2) | 18 (2) | 18 (2) | HP |
| Hypothetical protein BP2534 | NP_881161 | BP2534 | HP | 58 | U | 25 (8) | 18 (6) | 18 (6) | 17 (7) | HP/metabolism |
| Hypothetical protein BP2535 | NP_881162 | BP2535 | HP | 43 | CM | 34 (8) | 34 (8) | 14 (4) | 29 (7) | HP/metabolism |
| Hypothetical protein BP2661^ | NP_881275 | BP2661 | HP | 32 | U | 10 (2) | 5 (1) | 10 (2) | 10 (2) | HP/transport |
| Hypothetical protein BP2717 | NP_881325 | BP2717 | HP | 40 | M/U | 29 (5) | 14 (3) | 5 (2) | 28 (5) | HP |
| Hypothetical protein BP2936 | NP_881518 | BP2936 | HP | 37 | U | 23 (6) | 17 (4) | 36 (9) | 33 (8) | HP |
| Hypothetical protein BP3467 | NP_881990 | BP3467 | HP | 92 | U | 15 (7) | 15 (7) | 10 (5) | 14 (7) | HP/membrane biogenesis |
| Hypothetical protein BP3521 | NP_882036 | BP3651 | HP | 61 | C | 7 (3) | 6 (2) | 5 (2) | 7 (3) | HP/membrane biogenesis |
| Hypothetical protein BP3559 | NP_882072 | BP3559 | HP | 39 | M/U | 30 (6) | 32 (6) | 29 (6) | 20 (4) | HP/cell division |
| Hypothetical protein BP3651 | NP_882159 | BP3651 | HP | 77 | U | 7 (3) | 7 (3) | 9 (4) | 8 (3) | HP/membrane biogenesis |
| Hypothetical protein BP3689 (LysM domain/BON SFP) | NP_882194 | BP3689 | HP | 20 | U | 24 (3) | 22 (3) | 22 (3) | 22 (3) | HP/cell wall degradation |
| Hypothetical protein BP3758^ | NP_882261 | BP3758 | HP | 29 | CM | 3 (1) | 5 (1) | 7 (2) | 7 (2) | HP/membrane transport |
| Hypothetical protein BP3819 | NP_882317 | BP3819 | HP | 27 | U | 12 (2) | 23 (3) | 19 (2) | 19 (2) | HP |
| l-lactate dehydrogenase | NP_879338 | BP0484 | ildD | 43 | C | 16 (4) | 19 (3) | 8 (2) | 28 (6) | Carbohydrate metabolism |
| Large-conductance mechanosensitive channel | NP_879158 | BP0278 | mscl | 17 | U | 25 (3) | 25 (3) | 13 (2) | 25 (3) | Transport |
| Lipoprotein^ | NP_881354 | BP2750 | HP | 24 | U | 13 (2) | 13 (2) | 11 (2) | 7 (1) | U |
| Lipoprotein | NP_879963 | BP1189 | U | 16 | U | 24 (2) | 39 (3) | 39 (3) | 25 (2) | U |
| Mce related protein | NP_882262 | BP3759 | U | 18 | U | 59 (4) | 35 (2) | 73 (5) | 45 (3) | Transport |
| Outer membrane lipoprotein | NP_881135 | BP2508 | omlA | 20 | OM | 34 (3) | 29 (3) | 40 (4) | 47 (5) | Transport |
| Outer membrane porin protein OmpQ | NP_881933 | BP3405 | ompQ | 40 | OM | 30 (8) | 27 (7) | 23 (6) | 29 (8) | Transport |
| Outer membrane porin protein precursor | NP_879650 | BP0840 | U | 42 | OM | 52 (12) | 43 (10) | 47 (10) | 43 (10) | Transport |
| Outer membrane protein A precursor | NP_879744 | BP0943 | ompA | 21 | OM | 44 (7) | 39 (5) | 39 (6) | 44 (6) | Transport |
| Outer membrane usher protein precursor | NP_880573 | BP1882 | fimC | 96 | OM | 10 (6) | 11 (6) | 13 (9) | 11 (7) | Membrane biogenesis |
| Penicillin-binding protein 1A | NP_882163 | BP3655 | U | 90 | E | 9 (3) | 11 (4) | 10 (4) | 11 (5) | Peptidoglycan metabolism |
| Putative peptidoglycan-associated lipoprotein | NP_881875 | BP3342 | U | 18 | OM | 59 (7) | 59 (6) | 67 (8) | 53 (7) | Transport |
| Peptidyl-prolyl cis-trans isomerase D | NP_880447 | BP1732 | ppiD | 7 | U | 45 (16) | 38 (13) | 38 (14) | 36 (12) | Protein folding |
| Pertactin | BAF35031 | U | prn | 101 | OM | 21 (13) | 17 (11) | 14 (8) | 15 (9) | Transport |
| Phosphoenolpyruvate synthase | NP_880178 | BP1436 | ppsA | 87 | C | 9 (6) | 3 (2) | 5 (3) | 3 (2) | Carbohydrate metabolism |
| Phosphoglucomutase/phosphomannomutase | NP_879859 | BP1075 | qlmM | 50 | C | 9 (3) | 6 (2) | 6 (2) | 6 (2) | Carbohydrate metabolism |
| Preprotein translocase subunit SecA | NP_881589 | BP3014 | secA | 100 | CM | 10 (7) | 6 (4) | 2 (2) | 2 (2) | Membrane transport |
| Preprotein translocase subunit SecD | NP_879831 | BP1046 | secD | 69 | CM | 28 (11) | 23 (10) | 29 (11) | 26 (10) | Membrane transport |
| Preprotein translocase subunit SecF | NP_879830 | BP1045 | secF | 34 | CM | 19 (3) | 19 (3) | 19 (3) | 19 (3) | Membrane transport |
| Preprotein translocase subunit SecG | NP_879617 | BP0802 | secG | 16 | U | 41 (2) | 41 (2) | 41 (2) | 41 (2) | Membrane transport |
| Putative ABC transporter | NP_880959 | BP2321 | Pu | 69 | CM | 16 (5) | 10 (3) | 15 (4) | 15 (4) | Transport |
| Putative ABC transporter ATP-binding subunit^ | NP_881029 | BP2397 | Pu | 70 | CM | 7 (2) | 8 (2) | 2 (1) | 5 (1) | Membrane transport |
| Putative ABC transporter ATP-binding subunit^ | NP_880660 | BP1986 | Pu | 70 | CM | 3 (1) | 11 (4) | 7 (2) | 7 (2) | Membrane transport |
| Putative ABC transporter ATP-binding subunit | NP_882260 | BP3757 | Pu | 31 | CM | 19 (3) | 29 (4) | 29 (4) | 23 (4) | Membrane transport |
| Putative ABC transporter ATP-binding subunit | NP_880723 | BP2057 | Pu | 46 | CM | 34 (7) | 28 (6) | 6 (2) | 24 (5) | Membrane transport |
| Putative amino acid ABC transporter ATP-binding protein | NP_882326 | BP3828 | Pu | 27 | CM | 20 (3) | 15 (2) | 20 (3) | 15 (2) | Membrane transport |
| Putative amino acid ABC transporter permease protein | NP_882328 | BP3830 | Pu | 44 | CM | 13 (3) | 8 (2) | 8 (2) | 8 (2) | Membrane transport |
| Putative bifunctional protein | NP_882247 | BP3744 | Pu | 43 | CM | 22 (6) | 17 (4) | 12 (3) | 17 (4) | Energy metabolism |
| Putative binding-protein-dependent transport permease | NP_881026 | BP2394 | Pu | 33 | CM | 5 (1) | 3 (1) | 7 (2) | 5 (1) | Membrane transport |
| Putative binding-protein-dependent transport protein | NP_881859 | BP3322 | Pu | 42 | P | 41 (7) | 27 (6) | 33 (6) | 26 (4) | Membrane transport |
| Putative cell division protein^ | NP_881100 | BP2473 | Pu | 87 | CM | 5 (2) | 2 (1) | 4 (1) | 4 (3) | Cell division |
| Putative chromosome partition protein | NP_882071 | BP3558 | Pu | 130 | C | 7 (6) | 11 (7) | 9 (6) | 8 (6) | Cell division |
| Putative dioxygenase | NP_880971 | BP2333 | Pu | 34 | CM | 16 (3) | 19 (4) | 11 (2) | 19 (4) | Amino acid metabolism |
| Putative efflux system inner membrane protein | NP_880738 | BP2075 | Pu | 49 | CM | 14 (4) | 18 (5) | 14 (4) | 14 (4) | Membrane transport |
| Putative efflux system transmembrane protein | NP_880739 | BP2076 | Pu | 118 | CM | 12 (9) | 5 (4) | 8 (7) | 10 (7) | Membrane transport |
| Putative exported solute binding protein | NP_881542 | BP2963 | Pu | 40 | U | 17 (4) | 10 (2) | 10 (2) | 13 (3) | Membrane transport |
| Putative extracellular solute-binding protein | NP_880657 | BP1983 | Pu | 82 | P | 6 (3) | 6 (3) | 6 (3) | 6 (3) | Membrane transport |
| Putative glycosyl transferase | NP_881785 | BP3238 | Pu | 34 | U | 20 (4) | 10 (2) | 12 (2) | 22 (4) | Metabolism |
| Putative inner membrane protein | NP_881862 | BP3326 | Pu | 26 | U | 27 (4) | 27 (4) | 28 (4) | 49 (6) | HP |
| Putative inner membrane protein translocase component YidC | NP_886531 | BBP4405 | Pu | 62 | CM | 9 (3) | 15 (4) | 12 (4) | 7 (3) | Membrane transport |
| Putative inner membrane-anchored protein | NP_880838 | BP2190 | Pu | 33 | U | 16 (4) | 20 (5) | 20 (4) | 15 (3) | HP |
| Putative integral membrane protein | NP_881049 | BP2420 | Pu | 41 | CM | 9 (2) | 12 (2) | 16 (4) | 7 (2) | Membrane transport |
| Putative l-lactate dehydrogenase | NP_879251 | BP0379 | Pu | 39 | C | 7 (2) | 18 (3) | 18 (3) | 21 (4) | Carbohydrate metabolism |
| Putative lipoprotein | NP_880735 | BP2072 | Pu | 22 | U | 24 (3) | 27 (3) | 24 (3) | 18 (2) | Putative transport |
| Putative lipoprotein | NP_881568 | BP2992 | Pu | 18 | OM | 59 (6) | 49 (5) | 53 (5) | 53 (5) | Putative transport |
| Putative lipoprotein | NP_882263 | BP3760 | Pu | 29 | OM | 13 (2) | 25 (4) | 16 (4) | 11 (3) | Putative transport |
| Putative lipoprotein | NP_880063 | BP1296 | Pu | 30 | U | 16 (3) | 12 (2) | 12 (2) | 12 (2) | U |
| Putative lipoprotein | NP_880303 | BP1296 | Pu | 41 | U | 26 (5) | 30 (6) | 22 (5) | 30 (7) | Putative membrane |
| Putative lipoprotein | NP_880710 | BP2043 | Pu | 24 | U | 26 (4) | 15 (2) | 23 (4) | 11 (2) | U |
| Putative membrane transport ATPase | NP_881330 | BP2722 | Pu | 86 | CM | 4 (2) | 4 (2) | 3 (2) | 5 (2) | Putative transport |
| Putative membrane transport protein | NP_881309 | BP2716 | Pu | 49 | U | 7 (2) | 7 (2) | 11 (3) | 7 (2) | Putative transport |
| Putative NADH dehydrogenase | NP_882010 | BP3491 | ndh | 48 | CM | 10 (3) | 6 (2) | 6 (2) | 6 (2) | Energy metabolism |
| Putative outer membrane (permeability) protein | NP_881865 | BP3329 | Pu | 87 | OM | 16 (7) | 9 (4) | 12 (6) | 10 (5) | Putative transport |
| Putative outer membrane ligand binding protein | NP_879893 | BP1112 | bipA | 144 | OM | 39 (28) | 15 (12) | 23 (17) | 13 (12) | Adherence |
| Putative peptidase | NP_880436 | BP1721 | Pu | 32 | OM | 22 (3) | 20 (3) | 20 (3) | 27 (4) | Protein degradation |
| Putative quinoprotein | NP_880844 | BP2196 | Pu | 42 | OM | 31 (6) | 29 (5) | 38 (7) | 20 (4) | Protein assembly |
| Putative secreted protein^ | NP_879832 | BP1047 | Pu | 13 | U | 32 (2) | 32 (2) | 32 (2) | 21 (1) | Membrane transport |
| Putative secretion system protein | NP_882292 | BP3793 | ptle | 26 | U | 14 (3) | 21 (4) | 17 (3) | 16 (3) | Putative transport |
| Putative secretion system protein | NP_882293 | BP3794 | ptlf | 30 | U | 11 (2) | 19 (3) | 19 (3) | 11 (2) | Putative transport |
| Putative secretion system protein | NP_882289 | BP3794 | ptlc | 91 | CM | 6 (3) | 9 (5) | 9 (5) | 11 (6) | Putative transport |
| Putative sugar transport protein^ | NP_881176 | BP2549 | Pu | 61 | CM | 5 (2) | 2 (1) | 5 (2) | 5 (2) | Putative transport |
| Putative sulfatase | NP_881701 | BP3136 | Pu | 72 | CM | 21 (8) | 10 (5) | 17 (8) | 17 (8) | Membrane biogenesis |
| Putative TolQ-like translocation protein | NP_881879 | BP3346 | Pu | 25 | CM | 22 (3) | 22 (3) | 23 (4) | 19 (3) | Membrane transport |
| Putative type III secretion protein | NP_880879 | BP2235 | Pu | 66 | CM | 8 (3) | 10 (3) | 12 (5) | 12 (4) | Membrane transport |
| Pyrroline-5-carboxylate reductase | NP_880048 | BP1280 | proC | 30 | C | 15 (3) | 15 (3) | 10 (2) | 11 (2) | amino acid metabolism |
| RecA | AAK85426 | U | recA | 31 | C | 36 (6) | 31 (5) | 28 (4) | 22 (4) | DNA processing |
| Ribonuclease E | NP_879331 | BP0475 | rne | 115 | C | 12 (8) | 8 (5) | 6 (4) | 8 (5) | RNA degradation |
| Ribonucleotide-diphosphate reductase subunit alpha | NP_881559 | BP2983 | nrdA | 107 | C | 10 (7) | 5 (3) | 3 (2) | 8 (5) | Nucleotide metabolism |
| RNA polymerase sigma 80 subunit | AAC45085 | BP1191 | rpoD | 81 | C | 9 (4) | 5 (2) | 6 (3) | 6 (2) | RNA processing |
| Rod shape-determining protein | NP_879246 | BP0374 | mreB | 38 | C | 16 (3) | 19 (4) | 8 (2) | 12 (3) | Cell morphology |
| SCO1/SenC family protein^ | NP_882237 | BP3734 | U | 22 | U | 17 (3) | 6 (1) | 10 (2) | 6 (1) | Transport |
| Serum resistance protein | NP_882013 | BP3494 | brkA | 111 | OM | 23 (14) | 27 (14) | 29 (17) | 38 (21) | Transport |
| Signal peptidase I | NP_881060 | BP2432 | lep | 32 | CM | 11 (2) | 19 (4) | 14 (3) | 12 (2) | Transport |
| Succinate dehydrogenase flavoprotein subunit^ | NP_880997 | BP2361 | sdhA | 65 | CM | 7 (3) | 7 (3) | 3 (1) | 11 (5) | Carbohydrate metabolism |
| Succinate dehydrogenase iron–sulfur protein^ | YP_785707 | BAV1185 | sdhB | 26 | CM | 4 (1) | 14 (4) | 9 (3) | 14 (3) | Carbohydrate metabolism |
| Succinyl-CoA synthetase subunit beta | NP_881168 | BP2541 | sucC | 42 | U | 14 (3) | 9 (2) | 9 (2) | 17 (4) | Carbohydrate metabolism |
| Surface antigen | NP_880169 | BP1427 | U | 86 | OM | 29 (15) | 29 (15) | 25 (13) | 35 (18) | Protein assembly |
| Tex | CAA64672 | BP1144 | tex | 87 | C | 5 (3) | 4 (2) | 3 (2) | 4 (2) | RNA processing |
| Thiol:disulfide interchange protein | NP_882154 | BP3646 | DsbA | 71 | CM | 15 (7) | 14 (6) | 14 (6) | 10 (4) | Membrane biogenesis |
| Threonine synthase | NP_881383 | BP2783 | thrC | 51 | C | 16 (5) | 31 (9) | 10 (3) | 17 (5) | Amino acid metabolism |
| TonB-dependent receptor for iron transport | NP_879666 | BP0856 | bfrD | 82 | OM | 34 (20) | 35 (21) | 29 (18) | 28 (15) | Membrane transport |
| Tracheal colonization factor precursor | NP_879974 | BP1201 | tcfA | 71 | OM | 12 (6) | 20 (8) | 19 (10) | 10 (6) | Membrane transport |
| Translocation protein TolB | NP_881876 | BP3343 | tolB | 48 | P | 13 (3) | 15 (4) | 21 (6) | 14 (4) | Transport |
| Trifunctional transcriptional regulator/proline DH/P-5-C DH | NP_881353 | BP2749 | putA | 140 | C | 11 (9) | 10 (8) | 14 (12) | 14 (12) | Amino acid metabolism |
| Twin argininte translocase protein A | NP_882278 | BP3777 | tatA | 8 | U | 35 (2) | 35 (2) | 35 (2) | 35 (2) | Membrane transport |
| Type II citrate synthase | NP_880994 | BP2358 | qltA | 48 | C | 11 (3) | 8 (2) | 10 (3) | 10 (3) | Carbohydrate metabolism |
| Ubiquinol oxidase polypeptide I | NP_881514 | BP2932 | cyoB | 72 | CM | 6 (3) | 6 (2) | 9 (4) | 6 (2) | Energy metabolism |
| Uridylate kinase | NP_880163 | BP1421 | pyrH | 26 | C | 13 (3) | 10 (2) | 23 (4) | 16 (3) | Nucleotide metabolism |
| Virulence factors transcription regulator | NP_880570 | BP1878 | bvgA | 23 | C | 43 (7) | 59 (9) | 52 (8) | 34 (5) | Signal transduction |
| DNA polymerase I | NP_880026 | BP1254 | polA | 99 | C | 5 (3) | 5 (3) | Nucleotide metabolism | ||
| Acetyl-CoA synthetase | NP_881040 | BP2409 | acsA | 72 | C | 6 (3) | 4 (2) | Carbohydrate metabolism | ||
| 2-Oxoglutarate dehydrogenase E1 component^ | NP_879903 | BP1124 | sucA | 106 | C | 2 (1) | 4 (2) | Metabolism | ||
| Probable Orn/Arg/Lys decarboxylase^ | NP_879079 | BP0190 | U | 83 | C | 3 (1) | 4 (2) | Amino acid metabolism | ||
| Autotransporter | NP_879378 | BP0529 | U | 246 | OM/E | 7 (2) | 5 (6) | 2 (2) | Membrane transport | |
| Cytochrome B^ | NP_879156 | BP0276 | petB | 51 | CM | 17 (5) | 3 (1) | 5 (2) | Energy metabolism | |
| DNA mismatch repair protein^ | NP_879129 | BP0244 | mutL | 69 | C | 4 (2) | 6 (2) | 5 (1) | DNA processing | |
| HP BP3084 | NP_881655 | BP3084 | HP | 41 | C | 18 (4) | 17 (4) | 10 (2) | HP | |
| Ubiquinol-cytochrome C reductase iron–sulfur subunit | NP_879157 | BP0277 | petA | 23 | CM | 24 (3) | 24 (3) | 19 (2) | Energy metabolism | |
| 30s ribosomal protein S20^ | NP_881377 | BP2773 | rspT | 90 | U | 14 (2) | 13 (1) | 14 (2) | Translation | |
| Putative membrane-bound transglycosylase^ | NP_881812 | BP3268 | Pu | 47 | OM | 4 (1) | 14 (4) | 16 (4) | Membrane biogenesis | |
| Putative transglycosylase^ | NP_881631 | BP3060 | Pu | 76 | P | 4 (2) | 4 (1) | 4 (1) | Membrane biogenesis | |
| Dermonecrotic toxin | NP_881965 | BP3439 | dnt | 161 | U | 5 (4) | 2 (2) | 4 (4) | Cell death | |
| Putative periplasmic solute-binding | NP_880224 | BP1487 | smoM | 40 | U | 8 (2) | 8 (2) | 16 (4) | Putative transport | |
| Serotype 3 fimbrial subunit | NP_880302 | BP1568 | fim3 | 22 | E | 20 (3) | 27 (4) | 15 (2) | Cell integrity | |
| CTP synthetase (synthase) | NP_881022 | BP2389 | pyrG | 61 | C | 5 (2) | Nucleotide metabolism | |||
| Cycolysin secretion protein | NP_879580 | BP0762 | cyaD | 48 | CM | 12 (3) | Membrane transport | |||
| HP BP1123 | NP_879902 | BP1123 | HP | 75 | U | 11 (5) | HP | |||
| Putative ketopantoate reductase | NP_880110 | BP1360 | Pu | 34 | C | 10 (2) | HP/metabolism | |||
| Putative transcriptional regulation protein | NP_881198 | BP2571 | Pu | 15 | U | 27 (2) | HP/RNA processing | |||
| Serotype 2 fimbrial subunit | P05788 | BP1119 | fim2 | 23 | E | 21 (2) | Cell integrity | |||
| Threonyl-tRNA-synthetase | NP_880233 | BP1497 | thrS | 71 | C | 8 (3) | Translation | |||
| ABC transport protein, ATP-binding component | NP_881414 | BP2816 | metN | 40 | CM | 10 (2) | Membrane transport | |||
| HP BP3441 | NP_881967 | BP3441 | HP | 36 | M/U | 19 (3) | HP | |||
| BpH2 (novel histone — 18323) | AAB40156 | U | bph2 | 16 | U | 19 (2) | DNA processing | |||
| Putative heme receptor | NP_879314 | BP0456 | hemC | 82 | OM | 6 (3) | Membrane transport | |||
| Trigger factor | NP_880485 | BP1774 | tig | 48 | U | 8 (2) | Cell division | |||
| Exopolyphosphatase | NP_879857 | BP1073 | ppx | 55 | C | 4 (2) | Nucleotide metabolism | |||
| HP BP2486 | NP_881113 | BP2486 | HP | 45 | CM | 8 (2) | HP | |||
| HP BP3002 | NP_881577 | BP3002 | HP | 69 | C | 6 (2) | HP | |||
Numerical values are based on a greater that 95% Scaffold protein identification probability**.
Abbreviations: T — Tohama I, C — C056, D — D946 and F — F656.
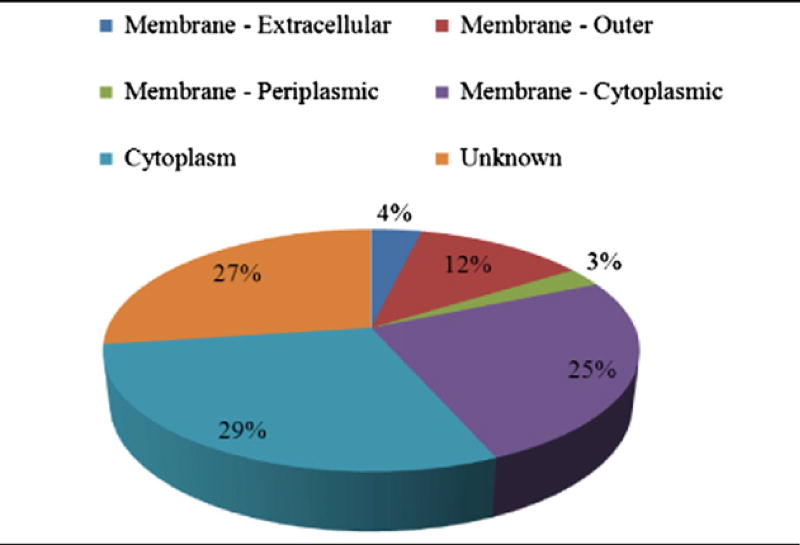
PSORTb subcellular scores were used to predict and localize the identified EMF proteins.
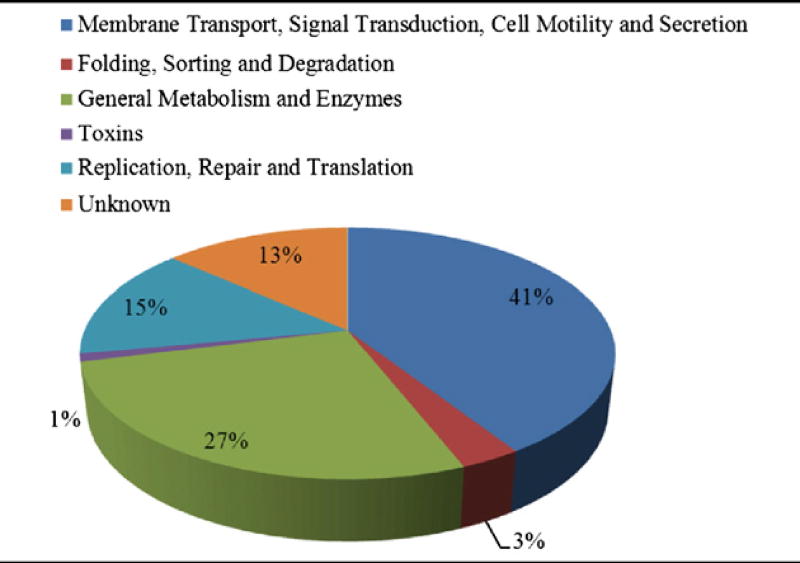
KEGG identifiers were employed to assign functions to each of the identified EMF proteins.
NCBI Gi accession numbers were used to further compile information such as gene identification number, subcellular localization (PSORTb) and protein function (KEGG identifiers). Values noted represent percent amino acid coverage of the identified EMF protein. (Number of unique peptides detected in parenthesis.).
indicates that identified proteins only had one unique detected tryptic peptide. No numerical value noted indicates that the protein was not identified in the strain.
Abbreviations: T — Tohama I, C — C056, D — D946 and F — F656; OM — outer membrane, CM — cytoplasmic membrane, C — cytoplasm, P — periplasm, E — extracellular, M — membrane, U — unknown, Pu — putative, HP — hypothetical protein; DH (dehydrogenase), P-5-C-DH (pyrroline-5-carboxylate DH), SFP (superfamily protein), 5-meta (5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase); BBP — B. parapertussis, BB — B. bronchiseptica, BAV – B. avium.