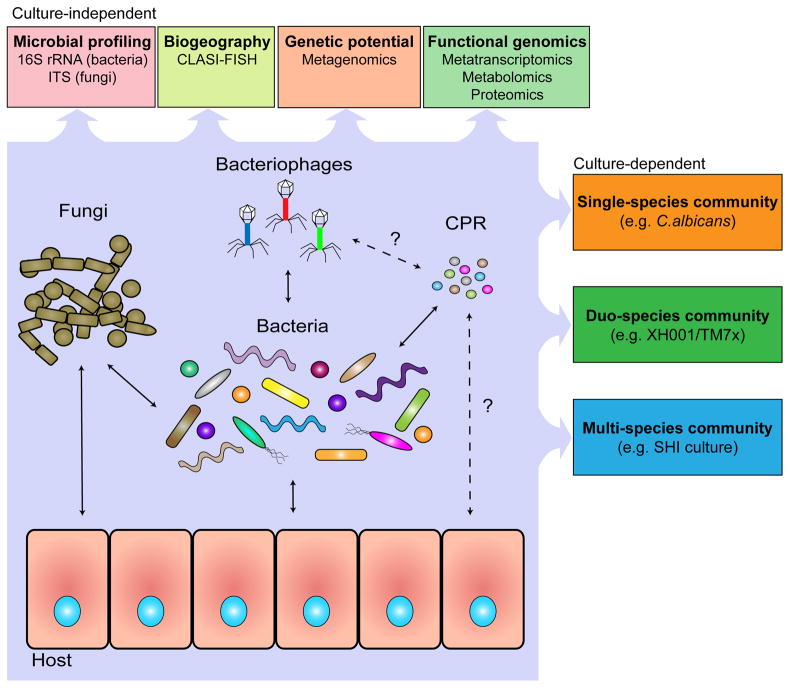
Figure 1: Key Figure. A systemic approach to the investigation of human oral microbiome.
A holistic understanding of human microbiome requires a systemic approach to better study the interactions within and between different oral microbial groups (bacteria, CPR, fungi and virus), their impact on microbial physiology, community ecology, and host homeostasis. State-of-the-art, culture-depended methods, coupled with traditional culture-dependent approaches allow researchers to better identify the characteristics of the microbiome, such as the prevalence and biogeography of all species present, as well as the interactions and metabolic pathways critical to the community and the effects of the community on the host. The application of these approaches has revealed a highly complex oral microbial community with sophisticated and dynamic interspecies interactions, which shape and define the community structure as well as functions. Within the oral microbiome, bacterial-bacterial, bacterial-fungal and bacterial-phage interactions and their impact on host have been extensively documented (solid arrows). Meanwhile, the improved cultivation approach, together with crucial genetic information obtained using culture-independent method resulted in the discovery of distinct bacterial-CPR interactions which could potentially prove to be common in host-associated microbiota (dotted arrows). Furthermore, culture-independent metagenomic analysis also suggested putative interspecies interactions, such as the presence of phage specifically targeting CPR members and direct interaction between CPR and the host (dashed arrows).