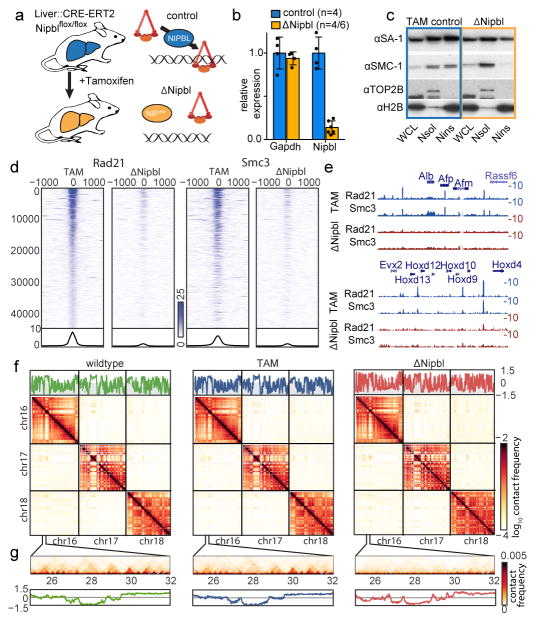
Figure 1. Overview of the experimental design.
(a) S We deleted a conditional allele of Nipbl (Extended Data Fig. 2) in adult hepatocytes using a liver-specific driver for the CRE-ERT2 fusion protein after injection of Tamoxifen. In absence of Nipbl, cohesin (represented by a triangular ring) is not loaded on chromatin. (b) Expression of Nipbl and Gapdh (control) by RT-qPCR in control (n=4) and ΔNipbl (n=6 for Nipbl; 4 for Gapdh) hepatocytes. Mean normalized gene expression (using Pgk1 as internal control, and WT expression level set as 1) is displayed as mean and s.d and compared using unpaired two-sided t-test (95% CI control=[0.7033–1.297]; mutant=[0.05731–0.2198] for Nipbl expression). (c) Western blots of hepatocyte protein extracts (WCL: whole cell lysate, Nsol (nuclear extract, soluble fraction) Nins (insoluble, chromatin fraction)) showed displacement of cohesin structural subunits (SA-1, SMC1) from the chromatin-bound fraction. H2B and TOP2B distribution serves as controls for loading and enrichment of two nuclear fractions. Experiment repeated on two biologically independent samples per condition. See Supplementary Data 1 for gel data source. (d) Stacked heatmaps of calibrated ChIP-seq signal (for Rad21 and SMC3) at WT Rad21 peaks ranked by fold change over input in the TAM control condition. (e) ChIP-seq tracks for Rad21 and SMC3 over representative genomic regions. (f–g) Hi-C maps at 20kb resolution of WT (left), TAM control (middle) and ΔNipbl cells (right). Top – cis compartment tracks. Middle – cis and trans contact maps of chr16–18. (g) An example of short-range Hi-C map at chr16:25–32Mb with compartment tracks.