ABSTRACT
Blood circulating cell-free DNA (cfDNA) is becoming popular in the search of promising predictive and prognostic biomarkers. Among these biomarkers, cfDNA methylation markers have especially gained considerable attention. A significant challenge in the utilization of cfDNA methylation markers is the limited amount of cfDNA available for analyses; reportedly, bisulfite conversion (BSC) reduce cfDNA amounts even further. Nevertheless, few efforts have focused on ensuring high cfDNA conversion efficiency and recovery after BSC. To compare cfDNA recovery of different BSC methods, we compared 12 different commercially available BSC kits. We tested whether DNA recovery was affected by the molecular weight and/or quantity of input DNA. We also tested BSC efficiency for each kit. We found that recovery varied for DNA fragments of different lengths: certain kits recovered short fragments better than others, and only 3 kits recovered DNA fragments of <100 bp well. In contrast, DNA input amount did not seem to affect DNA recovery: for quantities spanning between 820 and ∼25,000 genome equivalents per BSC, a linear relation was found between input and recovery amount. Overall, mean recovery ranged between 9 and 32%, with BSC efficiency of 97–99.9%. When plasma cfDNA was used as input for BSC, recovery varied from 22% for the poorest and 66% for the best performing kits, while conversion efficiency ranged from 96 to 100% among different kits. In conclusion, clear performance differences exist between commercially available BSC kits, both in terms of DNA recovery and conversion efficiency. The choice of BSC kit can substantially impact the amount of converted cfDNA available for downstream analysis, which is critical in a cfDNA methylation marker setting.
KEYWORDS: Biomarker, bisulfite conversion, circulating cell-free DNA, epigenetics, methylation, optimization
Introduction
Evaluation of blood circulating cell-free DNA (cfDNA) by looking at biomarkers is becoming increasingly popular for screening, diagnosis, and surveillance purposes in several diseases, including cancer,1,2 as cfDNA is released from both healthy and pathological tissue.3 cfDNA analyses have unique advantages compared with DNA analyses from ordinary tissue biopsies. First, with a routine, minimally invasive blood draw, DNA can be sampled as surrogate for the whole body, which is convenient in cases of inaccessible tissues that defy ordinary biopsy techniques. Second, implementing longitudinal monitoring of cfDNA is convenient, even more so when the tissue where the disease originated is unknown. Accordingly, liquid biopsies provide information regarding normal and disease-associated DNA, such as chromosomal alterations, sequence mutations, and epigenetic changes. Among the latter, aberrant cfDNA methylation patterns are currently gaining attention, particularly in cancer research,4,5 as they are known to arise early during cancer pathogenesis.6
It is well known that cfDNA-based methylation marker applications pose several technical challenges compared with traditional tissue-based marker ones. Foremost, cfDNA is short (with an average length of 147–167 bp),7,8 and pathological conditions such as cancer can aggravate fragmentation.9,10 Further, the amount of cfDNA in blood is limited, and only a small fraction of the isolated cfDNA originates from the pathological cells of interest (cell-free disease DNA, cdDNA). Therefore, highly sensitive biomarker detection methods are required, such as quantitative real-time PCR (qPCR), droplet digital PCR (ddPCR), and targeted next-generation sequencing (NGS), with persistent attention to cfDNA purification and recovery. Hence, several studies address how to ensure high recovery of ∼150 bp long double-stranded DNA from plasma.11–13
Importantly, a particular concern for the use of cfDNA-based methylation markers is the requirement of a bisulfite conversion (BSC) step before biomarker evaluation. This step enables discrimination of methylated and unmethylated cytosines. Unfortunately, a substantial amount of DNA is often lost during BSC due to chemical degradation of the DNA and sub-optimal post-conversion purification protocols.14,15 Reportedly, the loss is greater for low molecular weight (LMW)-DNA,14,16 such as cfDNA, than for high molecular weight (HMW)-DNA. Despite this, surprisingly few studies have addressed the challenge of achieving acceptable cfDNA recovery and conversion efficiency after cfDNA BSC.17–20
The aim of this study was to identify the BSC method best suited for cfDNA methylation marker research. We aimed for the greatest possible cfDNA recovery and highest conversion efficiency after BSC, to maximize the amount of cfDNA available for downstream methylation marker analysis. Our endpoints were post-BSC recovery, measured as the total amount of converted DNA in the elution; proportional DNA recovery, measured as the recovered converted DNA percentage of total DNA input; and BSC efficiency, measured as the proportion of un-converted DNA to the total amount of recovered DNA.
Results
We acquired 11 BSC kits from 10 different manufacturers. Furthermore, we tested an optimized custom-made protocol.20,21 An overview of different kit characteristics is available in Table 1; a flowchart providing an overview of the experiments performed can be found in Fig. S1.
Table 1.
Overview of bisulfite conversion methods*.
| Kit Name | Abbreviation | Input Volume, µl | Elution Volume, µl | Input DNA | Optimal Input DNA | Size | Recovery | Source | DNAProtection | Protocol Time (min) | 96-Format | Automation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MethylEasy Xceed | MethylEasy Xceed | 20 | 12 − 100 | 0.05 ng–5 µg | 0.05 ng–5 µg | — | >99.9% | Cells, tissue | Yes (Salmon sperm) | 90 | Yes (beads) | — |
| CpGenome™ Turbo Bisulfite Modification Kit | CpGenome Turbo | 10 | 25 + 25 | 0.5 ng–1 µg | 1 ng–1µg | — | — | Cells, tissue | No | 100 | No | — |
| EpiTect Plus Bisulfite Conversion | EpiTect Plus | 20-40 | 20 + 20 | 1 ng–2 µg | 1–2µg | — | — | All kinds | Yes (carrierRNA) | 370 | Yes | Yes |
| Bisulflash™ DNA Modification Kit | Bisulflash DNA Modification | 1–5 | 10 − 20 | 0.2 ng–1 µg | 50–200 ng | >100 bp, optimized for 250 bp | >90% | — | No | 60 | Yes | Yes (beads) |
| EpiJet Bisulfite Conversion Kit | EpiJet | 20 | 6 − 20 | 0.05 ng–2 µg | 200–500 ng | Optimized for >50 bp | — | — | No | 220 | No | Yes |
| Imprint DNA Modification Kit | Imprint DNA | 10 | 8 − 20 | 0.1 ng–1 µg | 50–200 ng | — | — | — | Yes (BSAsolution) | 155 | No | No |
| EpiMark Bisulfite Conversion Kit | Epimark | 10 | 20 + 20 | 50 ng–2 µg | 50 ng–2 µg | Optimized for >150 bp | — | — | No | 245 | No | No |
| EZ DNA Methylation Direct Kit | EZ DNA Methylation Direct | 20 | 10 | 50 pg - 2 µg | 200–500 ng | >17 bp | >80% | All kinds | No | 260 | Yes | Yes (beads) |
| Premium Bisulfite | Premium Bisulfite | 20 | 10 | 0.1 ng–2 µg | 200–500 ng | Optimized for bp>500 | >80% | — | No | 115 | No | Yes (column) |
| Bisulflash™ DNA Bisulfite Conversion Easy Kit | Bisulflash DNA Conversion | 2–15 | 10–20 | 100 ng–1 µg | 100 ng | >100 bp, optimized for 250 bp | >75% | Cells, tissue | No | 80 | Yes | No |
| innuCONVERT Bisulfite Body Fluids Kit | innu-CONVERT Bodyfluids | 50 | 20 − 100 | 0.5 ng–10 µg | — | Optimized for >100 bp | — | cfDNA in bodyfluids | Yes (carrierRNA) | 110 | No | — |
| Custommade Protocol | Custommade protocol | 35 | 35 | — | — | >17 bp | — | — | No | 50 | No | Yes |
*All information provided by manufacturers.
"-" Not available.
DNA recovery for 12 different BSC kits
Conversions of HMW- and LMW-DNA templates were done using the 12 kits. For the HMW-DNA template extracted from leucocytes, 100 ng of DNA were added to each reaction, as this quantity is within the range recommended by all manufacturers (Table 1).
Post-BSC DNA recovery, measured as the total amount of converted DNA in the elution, was assessed using BSC-dependent qPCR with the “MYOD1-assay," and revealed large inter-kit variation (Fig. 1A). The kits with the highest Post-BSC DNA recovery were the EpiJet, Epitect Plus, Premium Bisulfite, Imprint DNA, and EZ DNA Methylation Direct kits. For the LMW-DNA template (a 131 bp custom-designed PCR fragment), 1 ng of DNA was added per reaction. Recovery was evaluated using the BSC-dependent “BSC-assay.” Compared to the HMW-DNA template recovery, the inter-kit recovery variations were even more pronounced for the LMW-DNA template (Fig. 1A-B). Again, EZ DNA Methylation Direct, Premium Bisulfite, and Imprint DNA were among the kits with the highest post-BSC DNA recovery, but Bisulflash DNA Modification and innuCONVERT Bodyfluids also showed high DNA recovery for LMW-DNA template. The latter 2 kits were better ranked for LMW-DNA template than for HMW-DNA template, indicating that these kits might be better at recovering shorter than longer fragments.
Figure 1.
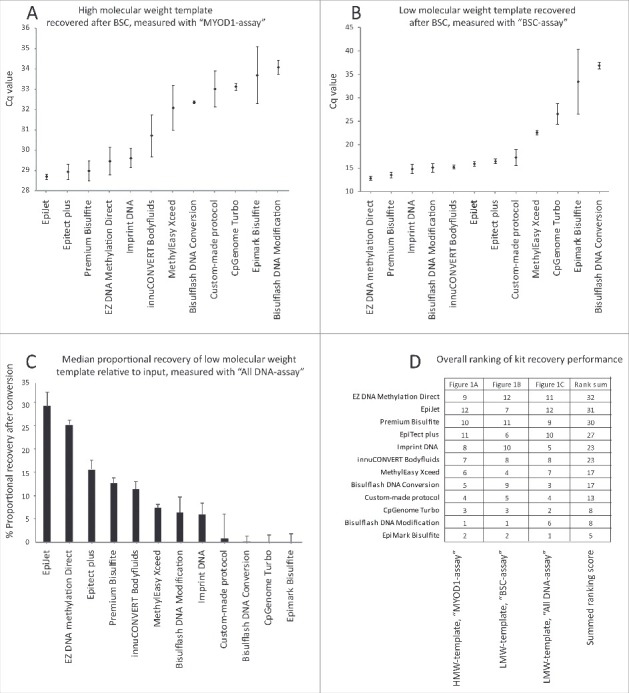
Impact of DNA molecular weight and fragment size on recovery after BSC. (A) Median HMW-DNA template quantity recovered after BSC assessed using the “MYOD1-assay” qPCR. Median Cq values of all technical replicates for 100 ng of input are shown. Whiskers represent median absolute deviation (MAD). (B) Median LMW-DNA template quantity recovered after BSC assessed by “BSC-assay” qPCR, Cq values of all technical replicates for 1 ng input are presented. Whiskers represent MAD. Please note that 1 ng input is below the recommended threshold for the Bisulflash DNA Conversion and Epimark kits. (C) Median proportional recovery of all replicates after BSC, analyzed by “All DNA-assay” for 1 ng input of LMW-DNA template (as percentage of input). Whiskers represent MAD. Bisulflash DNA Conversion and Bisulflash DNA Modification were only performed in 2 replicates. All other kits performed in 3 BSC replicates. (D) Recovery ranks for each kit and all 3 templates: HMW-DNA by “MYOD1-assay” (A); LMW-DNA by “BSC-assay” (B); and LMW-DNA by “All DNA-assay” (C). A score from 1–12 was given to each kit after each experiment, where ‘12’ corresponds to the kit with the lowest Cq value, i.e., the highest recovery, and ‘1’ corresponds to the kit with the highest Cq value, i.e., the lowest recovery. The 3 scores for each kit were added in to generate a final rank list. This rank was the basis for the selection of kits for further analyses.
In a BSC assay, DNA can be lost previous to recovery measurement in 3 different ways: i) It can be fragmented/degraded and, therefore, inaccessible for primer/probe annealing; ii) It can be lost during the purification process and, therefore, absent in the final elution and; iii) It may be un-converted if BSC efficiency is <100% and, therefore, undetected by quantification with a BSC-dependent PCR-assay (such as MYOD1-assay or BSC-assay). To investigate if the observed inter-kit DNA recovery differences were caused by differences during fragmentation/purification steps or due to different BSC efficiency, the “All DNA-assay,” which was designed to measure both BSC- and un-BSC LMW-DNA, was used to measure LMW-DNA template before and after BSC by each of the 12 kits, and proportional DNA recovery was assessed for each kit. A large inter-kit variation in DNA recovery was observed (Fig. 1C). The EZ DNA Methylation Direct, Epitect Plus, Premium Bisulfite, and innuCONVERT Bodyfluids kits were again top-ranked, as when the “BSC-assay” was used (Fig. 1B), indicating that inter-kit differences in DNA recovery were not caused by differences in bisulfite conversion efficiency, but rather by DNA fragmentation or purification loss. A ranking score from each of the 3 experiments can be seen in Fig. 1D.
DNA recovery depends on fragment length
Of the 12 kits initially tested, 5 kits consistently ranked high in DNA recovery: EpiJet, EZ DNA Methylation Direct, Epitect Plus, Premium Bisulfite, and Imprint DNA (Fig. 1D). These were therefore selected for additional testing.
To further examine how fragment size affected DNA recovery, 100 ng of a DNA Ladder-template (fragments 25 to 700 bp long) were bisulfite converted and the recovered DNA was visualized on a denaturing urea-polyacrylimide (PAGE) gel (Fig. 2 and Fig. S2). Densitometry of gel bands was then performed to quantify DNA recovery (Fig. 2B). Notably, recovery varied more for shorter cfDNA fragments (<200 bp) than for the longer fragments (≥200 bp). For example, the proportional recovery of 100 bp long DNA fragments spanned from <10% (EZ DNA Methylation Direct) to 80% (Premium Bisulfite), while recovery of 200 bp long DNA fragments spanned from 28% (Imprint DNA) to 66% (Epitect) (Fig. 2B). Recovery of 150 bp long DNA fragments (which are the approximate expected size of cfDNAs) was the highest when using the Premium Bisulfite, EZ DNA Methylation Direct, and EpiJet kits.
Figure 2.
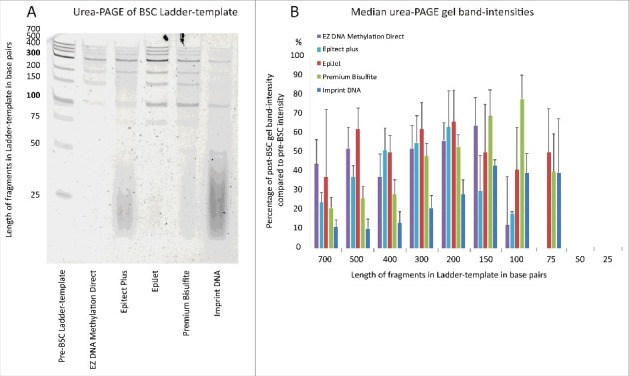
Fragment recovery assessment spanning DNA fragment sizes 25 to 700 bp. (A) Urea-PAGE of Ladder-template before and after BSC for 5 BSC kits. Dense smears are seen for Epitect Plus and Imprint DNA, where carrierRNA was added as per BSC protocol. All 5 kits have been run in triplicates. (B) Recovery of all fragments quantified by densitometry of urea-PAGE gel bands. Recovery = post-BSC/pre-BSC*100. Median band intensity values based on analysis of 3 independent gels are shown (see Fig. S3). Whiskers represent MAD.
Proportional recovery of cell-free-like-DNA is uniform across a wide range of DNA inputs
To examine how DNA input and DNA recovery were related, 5 Pool-templates with increasing quantities of sonicated DNA (average size ∼140 bp) were generated. The genome equivalents (GE) before BSC were 126 (0.4 ng) in Pool 1; 820 (2.7 ng) in Pool 2; 3,320 (11.0 ng) in Pool 3; 13,920 (45.9 ng) in Pool 4; and 24,889 (82.1 ng) in Pool 5. Genome equivalents were measured using the “Chr3-assay” by ddPCR. These DNA amounts span the most commonly observed cfDNA yields after purification from 1 mL of plasma. However, in a clinical setting, only the minor cdDNA sub-fraction of cfDNA is of interest, and this was mimicked by spiking an additional 500 copies of LMW-DNA template in the Pool-templates before BSC. This template mix enabled us to analyze if a cfDNA sub-fraction of interest (cdDNA mimicked by LMW-DNA template) was uniformly recovered across different inputs. Hereafter, Pool-templates were bisulfite converted using the 5 top-ranked kits (Fig. 1D), and DNA recovery was quantified using the “MYOD1-assay” for ddPCR (Fig. 3A). For Pool 1, with only 126 GE input, all 5 kits yielded insufficient copy numbers of bisulfite converted DNA to pass the limit of detection (LoD) of the “MYOD1-assay." Consequently, Pool 1 was excluded from this analysis. For the remaining Pool-templates, a linear relation was observed between DNA input and recovery in the range of 820 to ∼25,000 GE for all kits (Fig. S2). Proportional recovery was uniform across all DNA input amounts for individual kits, but overall mean DNA recovery varied: 9% (Imprint DNA), 21.5% (Epitect plus), 21.7% (EpiJet), 26.3% (Premium Bisulfite), and 32.5% (EZ DNA Methylation Direct) (Fig. 3A).
Figure 3.
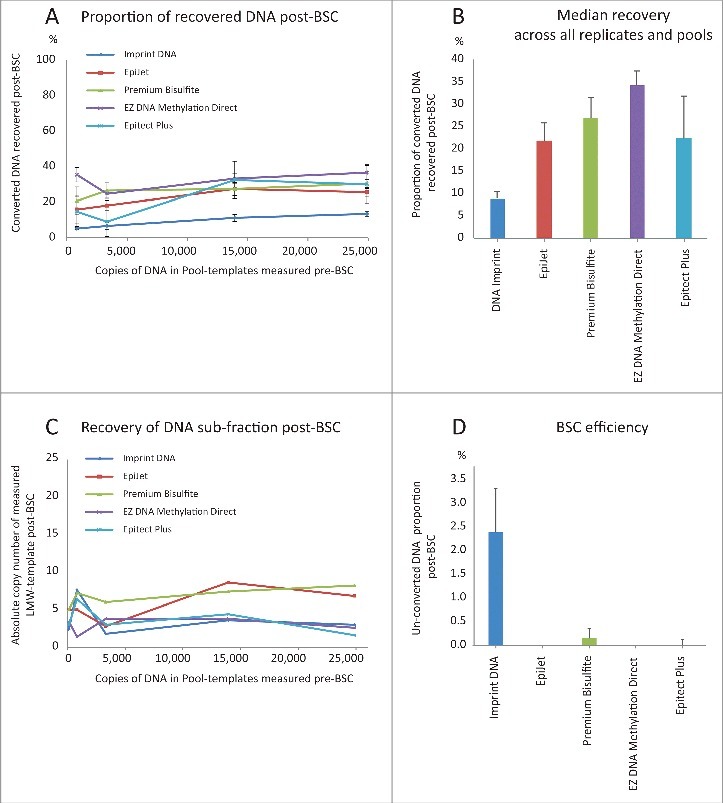
Impact of DNA input quantity on DNA recovery after BSC. (A) Median recovery (post-BSC/pre-BSC) for 4 different input quantities using Pool-templates 2–5 for all 5 kits. Whiskers represent MAD. Pool-template 1 was excluded from analysis, as measured DNA copy numbers in all reactions were below LoD for the “MYOD1-assay". One fifth of the BSC elusions were used as PCR input. (B) Median recovery for each kit across all technical replicates and all 5 pools. Whiskers represent MAD. (C) Median number of LMW-DNA template copies recovered by 5 different kits, using 5 different Pool-templates, and measured by the “BSC-assay". One fifth of BSC elusions were used as input for PCR. (D) Median percentage of un-converted DNA after BSC. Whiskers represent MAD. Data for all replicates of Pool-template 4 are shown. Similar data were obtained using all replicates of Pool-template 5 (not shown). Un-converted DNA was measured by the “Chr3-assay". One fifth of BSC DNA was used as PCR input.
To examine if the sub-fraction of LMW-DNA template recovered was independent of BSC input, the “BSC-assay” was subsequently applied to the Pool-templates (Fig. 3B and Fig. S2). Again, DNA recovery was uniform across all input quantities, indicating that the recovery of cdDNA was not affected by cfDNA input amount in this range.
Decreased bisulfite conversion efficiency may impair methylation marker performance
Clearly, BSC results depend on template recovery, but it is also imperative to simultaneously address the issue of BSC efficiency. To secure specificity of hypermethylated biomarkers, the efficiency of BSC must be close to 100%. If unmethylated cytosines are not converted to uraciles, the possibility of false positive results arises. Therefore, the proportion of fully un-converted DNA in the converted Pool-templates was estimated by the “Chr3-assay” using ddPCR for all Pool 4 replicates (Fig. 3C): Imprint DNA had >2% un-converted DNA in all replicates; Premium Bisulfite and Epitect Plus had 0–0.6% un-converted DNA; and EpiJet and EZ DNA Methylation Direct had 0%. Similar results were found for Pool 5 replicates (data not shown). Evidently, an internal BSC efficiency control must be part of any protocol when looking for aberrant methylation patterns in cfDNA.
Conversion of plasma cfDNA
Next, DNA recovery was tested on plasma cfDNA from 14 healthy individuals using the 5 top-ranked kits and the MethylEasy Xceed kit (Fig. 4A-E). The latter kit was included to address how a kit with suboptimal performance on artificial DNA samples performed on genuine cfDNA. cfDNA input quantities were estimated with the “Chr3-assay," and bisulfite converted DNA recovery was estimated with the “MYOD1-assay” in duplex with “Chr3-assay;" the former amplified all bisulfite converted cfDNA, the latter amplified only fully un-converted cfDNA. Recovery ranged from 10% (MethylEasy Xceed) to 66% (EZ DNA Methylation Direct) and BSC efficiency were between 88% (MethylEasy Xceed) and 100% (Premium Bisulfite). Correlation between pre- and post-BSC measurements was higher for the kits with the highest converted DNA recovery (R2 = 0.88 for EZ DNA Methylation Direct and R2 = 0.26 for MethylEasy Xceed). There were un-converted DNA remnants in at least one sample for all kits except Premium Bisulfite, which had a BSC efficiency of 100% (Fig. 4A-E).
Figure 4.
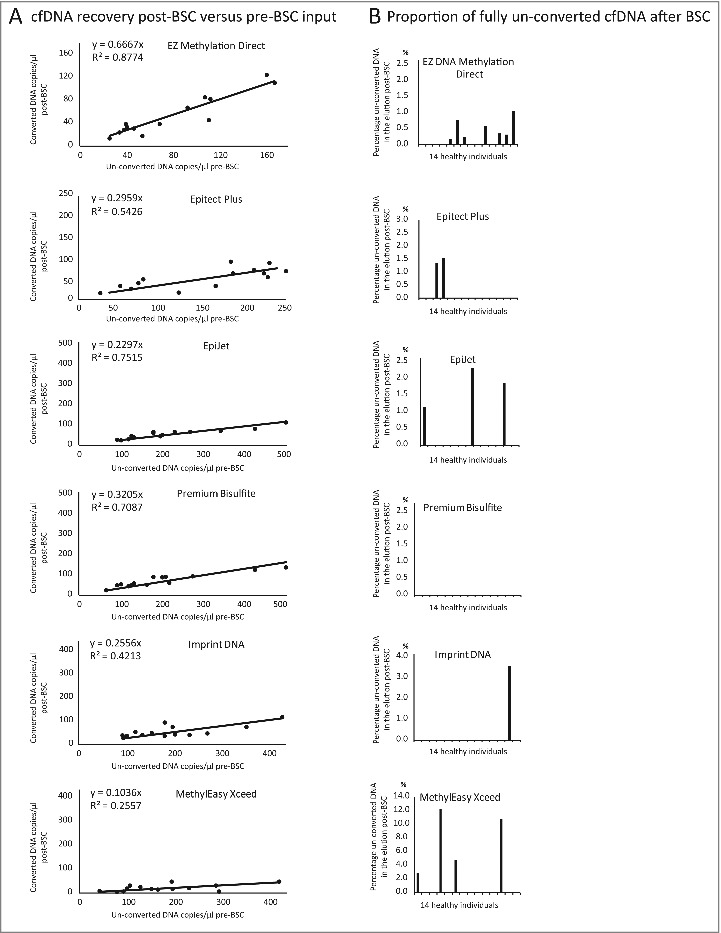
Relationship between plasma cfDNA input and post-BSC DNA recovery. (A) Post-BSC cfDNA recovery vs. pre-BSC cfDNA input quantity (measured using ”Chr3-assay” and “MYOD1-assay”) for 6 BSC kits. (B) BSC kit efficiency displayed as percentage of fully un-converted cfDNA in the total converted cfDNA after BSC.
Discussion
The aim of this study was to identify the best cfDNA BSC method. We tested 12 kits, including 11 commercial kits, by using 3 orthogonal methods (qPCR, urea-PAGE, and ddPCR) and 5 different DNA templates. It is well established that cfDNA is much shorter than genomic DNA from cell cultures or tissue; however, cfDNA size distribution is still disputed.2 Therefore, it is not possible to use artificial DNA of only one size to mimic cfDNA. Here, we used DNA fragment sizes ranging from 25 bp to several kb, with a clear focus on sizes 130–300 bp (LMW-DNA template), as most cfDNA is thought to be distributed around an average of 147–167 bp,2 with a significant fraction being even shorter.8,9,22 Also, a recent report suggests that disease-relevant circulating DNA species, such as ctDNA, may be even shorter (132–145 bp) than healthy cfDNA.10 Moreover, as we expected BSC recovery to decrease with shorter DNA fragments,18 we designed our LMW-DNA template to be in the lower end of the cfDNA size spectrum (131 bp) to avoid overestimation of BSC DNA recovery and ensure that our results would be useful to ctDNA researchers.
For cf-like-DNA, we generally found that only a small fraction of the input was recovered after BSC (9–32%), which is in agreement with previously published DNA recovery (2.7 to 86% for FFPE-DNA, cfDNA, and short PCR-produced fragments).17–19 In contrast, DNA recovery reported by most BSC kit manufacturers is considerably higher (>80%). A possible explanation for this discrepancy could be the use of different DNA quantification methods: when using PCR, only the DNA available for amplification is quantified; several manufacturers use a Qubit fluorometer, which quantifies all DNA.
DNA fragmentation and degradation after BSC are well-investigated causes for low input DNA recovery.15,23,24 As BSC kits are optimized for large, HMW-DNA inputs (typically >100 ng), this might further aggravate the degradation of small, LMW-DNA inputs. Among the kits tested here, innuCONVERT Bodyfluids was the sole kit specifically aimed at cfDNA. However, at present, it is unknown to what extent BSC-induced fragmentation is responsible for the loss of input DNA observed after BSC of cfDNA. The observed loss may occur simply due to small cfDNA fragments poor recovery by the DNA purification method following BSC. In our study, replicate urea-PAGE gels showed that small fragments (<100–150 bp) were more frequently lost compared with larger fragments (>300). Fragments shorter than 100 bp were only retained by 3 of 5 kits; fragments of this size may be of particular relevance when detecting very small cfDNA species, e.g., by direct sequencing.8 Whether small DNA fragments were lost due to chemical degradation or purification inefficiency was not determined by our experiments; however, Munson et al. suggested that especially the latter is crucial to small DNA fragment recovery.14 Although not formally tested here, the recovery of short DNA fragments (<100 bp) may enhance cfDNA detection. In line with this, we showed that some kits more efficiently recovered LMW-DNA templates than HMW-DNA templates, as the kits rank order shifted when one template replaced the other (e.g., innuCONVERT, Fig. 1A-B). It follows that cfDNA recovery might be increased by optimization of buffers and spin columns/beads toward retention of small DNA fragments.
As purified cfDNA yields vary greatly, it is important to know how BSC DNA input and recovery are related. Our results suggested that a lower input threshold exist. Below this amount, neither kit seemed able to recover sufficient DNA fragments for reliable detection; above it, DNA recovery was similar across the tested input range, although it varied between kits (9–32%) (Fig. 3A-B). This indicates that input quantity barely affected DNA recovery above a certain threshold, and our analysis estimated this threshold to lie below 820 GE. It is also important to keep in mind that when 820 GE are converted, DNA recovery from 9–32% will leave only 74–270 GE available for post-BSC analyses. If one presumes that sub-fractional cdDNA constitutes 1% or less of the cfDNA, then even with 820 GE input, inadequately few target DNA molecules would be recovered for downstream analyses. Consequently, for most practical purposes, a BSC DNA input well above 820 GE will be needed. Hence, the establishment of a lower DNA input amount detection threshold is of importance in planning and conducting cfDNA methylation marker studies, as it is critical to ensure that sufficient plasma volumes are collected to obtain a cfDNA amount that is well above this threshold.
Besides high cfDNA recovery, high BSC efficacy is pivotal: incomplete conversion of unmethylated cytosines may compromise methylation assay specificity and sensitivity. Especially in ‘rare-event’ analyses, supreme BSC efficacy is necessary as even very small remnants of non-converted DNA (<1%) may be more abundant than the pursued converted cdDNA. Generally, commercial BSC kits report bisulfite conversion efficiency of 90–100%. In agreement, we found most conversion rates varied between 99–100%. A small intra-kit variation is also likely due to technical differences in either the conversion process or in the ddPCR reactions. Nevertheless, we do recommend biomarker designs that distinguish fully bisulfite converted DNA from un-/hemi-converted DNA, and an integrated BSC efficiency control in the data analysis protocol. This will help ensure sufficient specificity and sensitivity in subsequent analyses.
In the presented experiments, DNA recovery using the same BSC kit differed (Figs. 1-4). This could partly be explained by differences in DNA template length, nucleotide complexity, and template amount. HMW-DNA template was genomic DNA, which is much longer and was used in higher amounts (100 ng) than the other templates. The LMW-DNA template was artificially generated, has low nucleotide complexity, and a fixed size of 131 bp. The Ladder-template has fixed sizes as well, whereas the sonicated Pool-templates were distributed around a maximum size of 140 bp. Moreover, methods to quantify DNA recovery differed (different PCR assays and urea-PAGE). However, there is coherence in DNA recovery across the experiments. For example, the EZ Methylation Direct kit recovered 0% of 75-bp fragments (Fig. 2). Recovery increased to 10% for 100-bp fragments (Fig. 2); 25.8% for 131-bp fragments (Fig. 1); 32% for 140-bp fragments (Fig. 4); and 80% for 150-bp fragment (Fig. 2).
Some of the limitations of our study include that, except in the case of LMW-DNA template, we compared template quantities before and after BSC using different PCR assays. Ideally, a cytosine-free assay should be used. The comparison of 2 different cytosine-containing assays is biased by the use of different PCR amplification conditions and efficiency. In addition, template regions can vary in number and primer/probe annealing accessibility, and, theoretically, BSC efficiency could be impacted by regional differences. However, as all kits are evaluated using the same PCR assays, this bias applies equally to all kits, and will not impact the relative ranking among the kits. Also, the recovery of LMW-DNA template was evaluated by amplifying a cytosine-free region using the “All DNA-assay” (Fig. 1C and Fig. S6) and kit performances rank overall similarly when evaluating recovery using 2 different PCR assays (Fig. 4A).
Another limitation of this study is that only 5 of the 12 kits were examined in all experiments. These 5 kits were selected based on their top ranking in the initial experiments using HMW- and LMW-DNA templates (Fig. 1D). DNA recovery and stable performance of the selected kids are supported by other studies17–20 and were corroborated when other types of templates were tested, including genuine cfDNA (Figs. 3 and 4).
In conclusion, this study showed that, for most kits, the recovery of converted DNA was affected by input DNA fragment length, but not by input DNA quantity. Generally, BSC efficiency was acceptably high, although un-converted DNA remnants sporadically were found for most kits. The study also showed that there was substantial inter-kit variation in the ability to recover cfDNA-sized fragments. Consequently, the choice of BSC kit may significantly impact the quantity of cfDNA available for downstream analyses. We recommend using one of the 5 top-ranked kits, although we also acknowledge that kit price and associated workload are of importance.
Materials and methods
DNA templates
Five different types of DNA templates were used for BSC. These enabled us to evaluate how DNA input quantity and DNA molecular weight impacted BSC recovery:
HMW-DNA template: DNA template purified from peripheral blood leukocytes. DNA was purified using the QiaSymphony DSP DNA Mini Kit (Cat. No. 937236, Qiagen), according to the manufacturer's protocol, and quantified using the Qubit 3.0 Fluorometer (Thermo Fischer Scientific).
LMW-DNA template: Artificial 131-bp long DNA fragment, mimicking ctDNA in size. The fragment was designed to consist of alternating cytosine-free and cytosine-containing stretches of DNA, thereby enabling the use of 2 different PCR assays with distinct properties (Fig. S4). The “BSC-assay” is capable of amplifying the LMW-DNA template only if bisulfite converted. This assay is used to estimate the recovery of converted template after BSC (Fig. S5). The “All DNA-assay” amplifies both converted and un-converted LMW-DNA template and was used to estimate the proportional template recovery after BSC compared with template input (Fig. S6). Large quantities of the LMW-DNA template were synthesized by PCR (Details in Fig. S4). PCR was performed using HiFi Polymerase (Cat. No. 11732641001, Roche) as follows: 1x HiFi-buffer, 1.95 U HiFi polymerase, and 0.2 mM dNTPmix were mixed with 0.2 mM forward- and 0.2 mM reverse-primer; RNAse-free water was added to a final volume of 50 µl. The mix was incubated on a thermal cycler under the following conditions: 95°C for 2 min, 10 cycles at 95°C for 30 sec, 56°C for 30 sec, and 72°C for 30 sec. The primers used for synthesis of the LMW-DNA template are listed in Table S1. To confirm the expected nucleotide sequence of the template, Sanger-sequencing was performed before and after BSC (data not shown). The LMW-DNA template was quantified using a Qubit 3.0 fluorometer.
Ladder-template: A DNA ladder (Cat. No. SM1191, Thermo Fisher Scientific) with fragments of 25, 50, 75, 100, 150, 200, 250, 300, 500, and 700 bp was used to analyze recovery of different fragment sizes.
Pool-templates: Five cfDNA-like pools with increasing DNA concentration (Pool-templates 1–5) were generated to reveal the correlation between DNA input and BSC recovery. To mimic cfDNA, leucocyte DNA was fragmented to ∼140 bp using a E220 Evolution Focused Ultrasonicator (Covaris, Inc.). Fragmentation was confirmed using a HT DNA Extended Range Labchip (Caliper Life Sciences) (Fig. S7). The 5 pools were constructed so that adding 2 µl of each individual Pool-template to a BSC reaction resulted in an estimated 100; 500; 2,000; 10,000; or 20,000 GE per reaction. In addition, each pool was spiked with LMW-DNA template, so that any given BSC additionally contained 500 copies of LMW-DNA template. To confirm the estimated Pool-template quantities (HMW- and LMW-DNA), these were measured using ddPCR before and after BSC.
cfDNA templates: cfDNA was purified from 8 mL of plasma from healthy individuals, as described previously,25 using the QiaSymphony Circulating Nucleic Acid Kit (Cat. No. 1017647; Qiagen) according to the manufacturer's instructions. Purified cfDNA was eluted in 60 µl and stored at -20°C for <1 month before use. cfDNA quantity, purity, and plasma purification efficiency were estimated by ddPCR before BSC, as described previously.26 The mean purified cfDNA quantity was 19,000 copies (62 ng) per individual, mean plasma purification efficiency was 97%, and no leucocyte remnants were measured in the elution. Plasma cfDNA was aliquoted and 6 selected BSC kits were applied to a total of 14 cfDNA samples each. Bisulfite conversions were performed manually. Post-BSC, the “MYOD1-assay“ and the “Chr3-assay” were run in duplex using ddPCR to quantify cfDNA recovery and BSC efficiency.
Bisulfite conversion
The different BSC kits were kindly provided by the manufacturers. For all kits, BSC was performed according to manufacturers’ protocol in triplicates. If DNA protection was recommended, carrierRNA was used. The workflow for each kit is presented in Fig. S8. To facilitate recovery comparison, template elusions were brought to equal volumes with nuclease-free water, before being stored at -20°C for <1 month.
PCR assays
MYOD1-assay: To quantify genomic, bisulfite converted DNA templates, we used a previously reported MethyLight assay, targeting chromosome 11.27,28
Chr3-assay: All un-converted genomic DNA templates were quantified using a previously reported TaqMan assay, targeting a locus on the short arm of chromosome 3.25 The Chr3-assay comprises 5 Gs in each primer and 7 Gs in the probe, which do not anneal to converted genomic DNA, where the paired cytosines will have been converted to uraciles. In silico analysis using BiSearch29 predicted, and ddPCR of several control samples (data not shown) confirmed, that the Chr3-assay did not amplify fully converted DNA.
BSC-assay: This assay was designed to target the cytosine-containing regions in the LMW-DNA template lagging strand. Primers only annealed to the template sequence if the cytosines were converted to uraciles.
All DNA-assay: The All DNA-assay targeted the cytosine-free regions of the LMW-DNA template leading strand. Cytosine-free sequences are unaltered by BSC; therefore, primers are able to amplify all LMW-DNA templates (converted and un-converted).
qPCR
qPCR experiments were conducted in house according to MIQE-guidelines30 (qMIQE checklist is shown in Table S2). For details about linearity, dynamic range, and PCR efficiency of qPCR assays, see Figs. S9 and S10.
qPCR was performed in technical triplicates on a ViiA7 Real Time PCR System (Life Technologies). The reaction mix was prepared using a Zephyr robot (Caliper Life Sciences) in MicroAmp® Optical 384-Well Reaction Plates (Cat. No. 4309849; Thermo Fisher Scientifc) using 2 µl of DNA template in a total PCR reaction volume of 5 µl. The qPCR reaction for the MYOD1-assay and Chr3-assay consisted of 0.4 mM forward primer, 0.4 mM reverse primer, 0.05 mM probe, and 1x of TaqMan Universal Master Mix (no UNG) (Cat. No. 4324018; Thermo Fisher Scientific) and 0.2 µl dNTPs. The qPCR reactions for the LMW-DNA template BSC-assay and the All DNA-assay contained 0.6 mM forward primer, 0.6 mM reverse primer, 0.1 mM probe, 1x TaqMan Universal Master Mix (no UNG), 0.1x TEMPase Hot Start (Cat. No. A220003; Ampliqon) and 0.2 µl dNTPa. Plates were sealed with MicroAmp® Optical Adhesive Film (Cat. No. 4311971; Thermo Fisher Scientific) and then amplified on a S1000 Thermal Cycler (BioRad) as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 sec, 56°C for 1 min. Results were analyzed using ViiA™ 7 Software, v1.2.3 (Applied Biosystems). Results are presented as median values with median absolute deviations; no outlier values were removed before analysis. Each PCR run included no-template controls, which were always blank.
ddPCR
ddPCR experiments were conducted according to MIQE-guidelines31 (dMIQE checklist is shown Table S3). For data output examples see Figs. S11 and S12. The reaction mix was manually prepared in a total 20-µl volume with 2 µl of template input, and consisted of 18 mM forward and reverse primer, 0.05 mM probe, 0.02x Supermix for probes (no UTP) (Cat. No. 186–3023; Bio-Rad). Droplets were generated on an automatic droplet generator QX200 AutoDG Droplet Digital PCR System (Bio-Rad) and analyzed in a S1000 Thermal Cycler using the following program: 95°C for 10 min, 45 cycles at 95°C for 30 sec, 56°C for 1 min and, finally, 98°C for 10 min. For the ”BSC-assay," 0.1 µl TEMPase Hot Start was further added to each reaction to ensure efficient amplification. Samples were analyzed on a QX100 Reader (Bio-Rad), and data were processed with Quantasoft v1.6.6 software (Bio-Rad). Partitions in each well were >12,000 droplets, with a mean of 16,000 for un-converted DNA, and a mean of 13,000 for converted DNA. Partition size was 1 nanoliter. No outliers were excluded and all data was Poisson-corrected. All PCR runs included positive, negative, and no-template controls. The MYOD1-assay and Chr3-assay were used either as single assays or in duplex with one another, as tests had shown that duplexing did not affect their performance. For each assay, 30 negative control samples and a varying number of positive control samples were profiled to establish the limit of blank (LoB) and limit of detection (LoD), as described previously.32 LoB for the MYOD1-assay was 11.9 copies/ddPCR well, LoD was 19.2 copies/ddPCR well. LoB for the Chr3-assay and BSC-assay was 0 copies/ddPCR well for both assays; their LoD was 3 copies/ddPCR-well32.
Polyacrylamide gel electrophoresis
To visualize recovery of the Ladder-template, urea-polyacrylamide gel electrophoresis (urea-PAGE) was performed. A denaturing, urea-based gel was used to avoid secondary DNA-folding of the single-stranded converted molecules. Polyacrylamide gels (12%) containing 7.5 M urea were cast with SequaGel - UreaGel (Cat. No. EC-833, National Diagnostics) and pre-run at 12 W for 1 h in 1x TBE. Samples (5 µl) were mixed with 20 µl urea loading-buffer (8 M urea, 2 mM Tris (pH 7.5), 20 mM EDTA, Bromophenol Blue (Cat. No. 57–13–6, 77–86–1, 60–00–4, and 115–39–9, respectively; Sigma-Aldrich). Samples were then denatured at 95°C for 60 sec and loaded into the gel. The samples were run for 30 min at 12 W and then stained with SYBR-gold (Cat. No. S11494, Thermo Fisher Scientific) in 1x TBE for 30 min. Bands were visualized with UV light. Band intensities were quantified by densitometry using Imagelab 4.1 Software (v. 2012; Bio-Rad).
Supplementary Material
Funding Statement
This work was supported by the Danish Council for Independent Research (11–105240), the Danish Council for Strategic Research (1309–00006B), the Novo Nordic Foundation (NNF14OC0012747), the Danish Cancer Research Foundation and the Danish Cancer Society (R133-A8520 and R40-A1965–11-S2).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors wish to thank the Danish Blood Donors for donating the blood samples and are grateful for the help provided by the dedicated staff at the Blood Bank at Aarhus University Hospital. We thank Poul Henning Madsen for critical reading of the manuscript and for sharing his know-how on bisulfite conversion.
References
- 1.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017; 31:172–9; PMID:28196593; https://doi.org/10.1016/j.ccell.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 2.Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17(4):223–38; PMID:28233803; https://doi.org/10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 3.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61:1659–65; PMID:11245480 [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6:224ra224; PMID:24553385; https://doi.org/10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orntoft MB, Nielsen HJ, Orntoft TF, Andersen CL, Danish Study Group on Early Detection of Colorectal C Performance of the colorectal cancer screening marker Sept9 is influenced by age, diabetes and arthritis: a nested case-control study. BMC Cancer 2015; 15:819; PMID:26514170; https://doi.org/10.1186/s12885-015-1832-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003; 3:253–66; PMID:12671664; https://doi.org/10.1038/nrc1045 [DOI] [PubMed] [Google Scholar]
- 7.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998; 17:89–97; PMID:9667526; https://doi.org/10.1097/00006676-199807000-00012 [DOI] [PubMed] [Google Scholar]
- 8.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its Tissues-Of-Origin. Cell 2016; 164:57–68; PMID:26771485; https://doi.org/10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, Gongora C, Thierry AR. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 2011; 6:e23418; PMID:21909401; https://doi.org/10.1371/journal.pone.0023418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J. Fragment length of circulating tumor DNA. PLoS Genet 2016; 12:e1006162; PMID:27428049; https://doi.org/10.1371/journal.pgen.1006162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA, Huggett JF. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014; 406:6499–512; PMID:24853859; https://doi.org/10.1007/s00216-014-7835-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013; 424:222–30; PMID:23727028; https://doi.org/10.1016/j.cca.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 13.Mauger F, Dulary C, Daviaud C, Deleuze JF, Tost J. Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem 2015; 407:6873–8; PMID:26123439; https://doi.org/10.1007/s00216-015-8846-4 [DOI] [PubMed] [Google Scholar]
- 14.Munson K, Clark J, Lamparska-Kupsik K, Smith SS. Recovery of bisulfite-converted genomic sequences in the methylation-sensitive QPCR. Nucleic Acids Res 2007; 35:2893–903; PMID:17439964; https://doi.org/10.1093/nar/gkm055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 2001; 29:E65–65; PMID:11433041; https://doi.org/10.1093/nar/29.13.e65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genereux DP, Johnson WC, Burden AF, Stoger R, Laird CD. Errors in the bisulfite conversion of DNA: modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res 2008; 36:e150; PMID:18984622; https://doi.org/10.1093/nar/gkn691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryzgonova O, Laktionov P, Skvortsova T, Bondar A, Morozkin E, Lebedeva A, Krause H, Miller K, Vlassov V. Efficacy of bisulfite modification and recovery of human genomic and circulating DNA using commercial kits. Eur J Mol Biol 2013; 1:1–8; https://doi.org/10.11648/j.ejmb.20130101.11 [Google Scholar]
- 18.Holmes EE, Jung M, Meller S, Leisse A, Sailer V, Zech J, Mengdehl M, Garbe LA, Uhl B, Kristiansen G, et al. . Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS One 2014; 9:e93933; PMID:24699908; https://doi.org/10.1371/journal.pone.0093933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leontiou CA, Hadjidaniel MD, Mina P, Antoniou P, Ioannides M, Patsalis PC. Bisulfite conversion of DNA: Performance comparison of different kits and methylation quantitation of epigenetic biomarkers that have the potential to be used in non-invasive prenatal testing. PLoS One 2015; 10:e0135058; PMID:26247357; https://doi.org/10.1371/journal.pone.0135058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedersen IS, Krarup HB, Thorlacius-Ussing O, Madsen PH. High recovery of cell-free methylated DNA based on a rapid bisulfite-treatment protocol. BMC Mol Biol 2012; 13:12; PMID:22448717; https://doi.org/10.1186/1471-2199-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayatsu H, Negishi K, Shiraishi M. Accelerated bisulfite-deamination of cytosine in the genomic sequencing procedure for DNA methylation analysis. Nucleic Acids Symp Ser (Oxf) 2004; 261–2; ISSN:17468272; PMID:17150578; https://doi.org/10.1093/nass/48.1.261 [DOI] [PubMed] [Google Scholar]
- 22.Andersen RF, Spindler KL, Brandslund I, Jakobsen A, Pallisgaard N. Improved sensitivity of circulating tumor DNA measurement using short PCR amplicons. Clin Chim Acta 2015; 439:97–101; PMID:25446878; https://doi.org/10.1016/j.cca.2014.10.011 [DOI] [PubMed] [Google Scholar]
- 23.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res 2007; 35:e29; PMID:17259213; https://doi.org/10.1093/nar/gkl1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka K, Okamoto A. Degradation of DNA by bisulfite treatment. Bioorg Med Chem Lett 2007; 17:1912–5; PMID:17276678; https://doi.org/10.1016/j.bmcl.2007.01.040 [DOI] [PubMed] [Google Scholar]
- 25.Reinert T, Scholer LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, et al. . Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016; 65:625–34; PMID:25654990; https://doi.org/10.1136/gutjnl-2014-308859 [DOI] [PubMed] [Google Scholar]
- 26.Pedersen SK, Symonds EL, Baker RT, Murray DH, McEvoy A, Van Doorn SC, Mundt MW, Cole SR, Gopalsamy G, Mangira D, et al. . Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer 2015; 15:654; PMID:26445409; https://doi.org/10.1186/s12885-015-1674-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeronimo C, Usadel H, Henrique R, Oliveira J, Lopes C, Nelson WG, Sidransky D. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst 2001; 93:1747–52; PMID:11717336; https://doi.org/10.1093/jnci/93.22.1747 [DOI] [PubMed] [Google Scholar]
- 28.Haldrup C, Mundbjerg K, Vestergaard EM, Lamy P, Wild P, Schulz WA, Arsov C, Visakorpi T, Borre M, Hoyer S, et al. . DNA Methylation signatures for prediction of biochemical recurrence after radical Prostatectomy of clinically localized prostate cancer. J Clin Oncol 2013; 31(26):3250–8; PMID:23918943; https://doi.org/10.1200/JCO.2012.47.1847 [DOI] [PubMed] [Google Scholar]
- 29.Aranyi T, Tusnady GE. BiSearch: ePCR tool for native or bisulfite-treated genomic template. Methods Mol Biol 2007; 402:385–402; PMID:17951807; https://doi.org/10.1007/978-1-59745-528-2_20 [DOI] [PubMed] [Google Scholar]
- 30.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. . The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55:611–22; PMID:19246619; https://doi.org/10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 31.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, et al. . The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin Chem 2013; 59:892–902; PMID:23570709; https://doi.org/10.1373/clinchem.2013.206375 [DOI] [PubMed] [Google Scholar]
- 32.Milbury CA, Zhong Q, Lin J, Williams M, Olson J, Link DR, Hutchison B. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif 2014; 1:8–22; PMID:27920993; https://doi.org/10.1016/j.bdq.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


