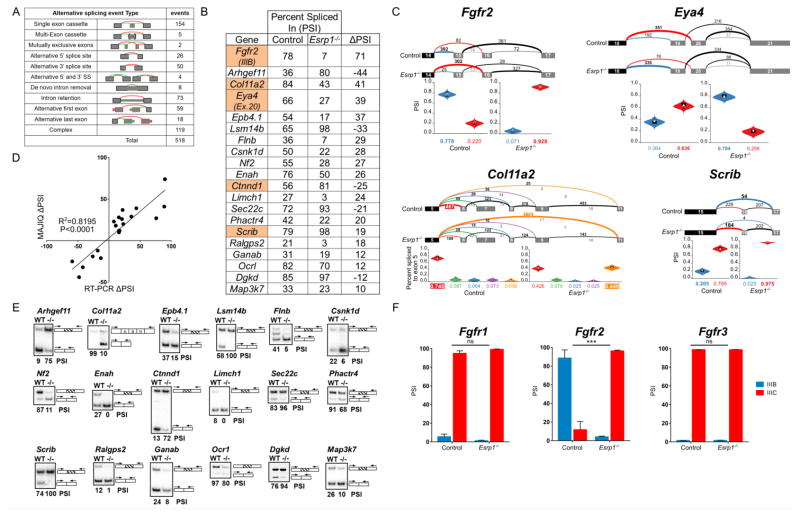
Figure 6. Alternative splicing is impaired in the cochlear epithelium of Esrp1−/− embryos.
(A) Number and type of alternative splicing switches identified by MAJIQ between Esrp1−/− and control embryos in the cochlear epithelium at E16.5 (ΔPSI > 10%). (B) PSI (percent spliced in) values for selected genes with significant splicing differences between Esrp1−/− and control embryos determined by MAJIQ and subsequently validated by RT-PCR. The highlighted genes have known roles in inner ear development and/or auditory function. (C) Voila views (Vaquero-Garcia et al., 2016) of Esrp1 dependent alternative splicing events for four genes associated with hearing loss showing examples of mutually exclusive (Fgfr2, Eya4), complex (Col11a2) and single (Scrib) exon cassettes. Numbered exons for each gene are represented as rectangles. Numbers above splicing events indicate RNA-seq read counts. Violin plots represent PSI estimates. (D) Correlation plot of alternative splicing switches predicted by MAJIQ and validated by RT-PCR. The Pearson’s product-moment correlation and the line of best fit (least squares polynomial) were computed in R (P<0.0001). (E) Radioactive RT-PCR validation of differential splicing events in the cochlear epithelium of Esrp1−/− and control littermates at E16.5. (F) Comparison of PSI values for Fgfr1-3, exon III-b (epithelial) and exon III-c (mesenchymal) in cochlear epithelium from Esrp1−/− and control embryos as determined by RT-PCR (multiple t-test comparison, P<0.0001). See also Table S1.