The meningococcal antigen typing system (MATS) is an enzyme-linked immunosorbent assay (ELISA)-based system that assesses the levels of expression and immune reactivity of the three recombinant MenB-4C antigens and, in conjunction with PorA variable 2 (VR2) sequencing, provides an estimate of the susceptibility of NmB isolates to killing by MenB-4C-induced antibodies. MATS assays or similar antigen phenotype analyses assume importance under conditions in which analyses of vaccine coverage predictions are not feasible with existing strategies, including large efficacy trials or functional antibody screening of an exhaustive strain panel. MATS screening of a panel of NmB U.S. isolates (n = 442) predicts high MenB-4C vaccine coverage in the United States.
KEYWORDS: MATS, MenB-4C, NHBA, NadA, Neisseria meningitidis, PorA, SBA, fHbp
ABSTRACT
Neisseria meningitidis is the most common cause of bacterial meningitis in children and young adults worldwide. A 4-component vaccine against N. meningitidis serogroup B (MenB) disease (MenB-4C [Bexsero]; GSK) combining factor H binding protein (fHBP), neisserial heparin binding protein (NHBA), neisserial adhesin A (NadA), and PorA-containing outer membrane vesicles was recently approved for use in the United States and other countries worldwide. Because the public health impact of MenB-4C in the United States is unclear, we used the meningococcal antigen typing system (MATS) to assess the strain coverage in a panel of strains representative of serogroup B (NmB) disease in the United States. MATS data correlate with killing in the human complement serum bactericidal assay (hSBA) and predict the susceptibility of NmB strains to killing in the hSBA, the accepted correlate of protection for MenB-4C vaccine. A panel of 442 NmB United States clinical isolates (collected in 2000 to 2008) whose data were down weighted with respect to the Oregon outbreak was selected from the Active Bacterial Core Surveillance (ABCs; CDC, Atlanta, GA) laboratory. MATS results examined to determine strain coverage were linked to multilocus sequence typing and antigen sequence data. MATS predicted that 91% (95% confidence interval [CI95], 72% to 96%) of the NmB strains causing disease in the United States would be covered by the MenB-4C vaccine, with the estimated coverage ranging from 88% to 97% by year with no detectable temporal trend. More than half of the covered strains could be targeted by two or more antigens. NHBA conferred coverage to 83% (CI95, 45% to 93%) of the strains, followed by factor H-binding protein (fHbp), which conferred coverage to 53% (CI95, 46% to 57%); PorA, which conferred coverage to 5.9%; and NadA, which conferred coverage to 2.5% (CI95, 1.1% to 5.2%). Two major clonal complexes (CC32 and CC41/44) had 99% strain coverage. The most frequent MATS phenotypes (39%) were fHbp and NHBA double positives. MATS predicts over 90% MenB-4C strain coverage in the United States, and the prediction is stable in time and consistent among bacterial genotypes.
IMPORTANCE The meningococcal antigen typing system (MATS) is an enzyme-linked immunosorbent assay (ELISA)-based system that assesses the levels of expression and immune reactivity of the three recombinant MenB-4C antigens and, in conjunction with PorA variable 2 (VR2) sequencing, provides an estimate of the susceptibility of NmB isolates to killing by MenB-4C-induced antibodies. MATS assays or similar antigen phenotype analyses assume importance under conditions in which analyses of vaccine coverage predictions are not feasible with existing strategies, including large efficacy trials or functional antibody screening of an exhaustive strain panel. MATS screening of a panel of NmB U.S. isolates (n = 442) predicts high MenB-4C vaccine coverage in the United States.
INTRODUCTION
Neisseria meningitidis, responsible for a global annual disease burden of ~50,000 deaths and as many long-term disabilities, is currently the most common cause of bacterial meningitis in children and young adults (1, 2). In the United States, serogroup B (NmB) strains cause approximately 30% of disease in all age groups, including adolescents, and >60% of cases in infants aged <1 year. Licensed conjugate vaccines have been successful in reducing the circulation and disease burden of serogroups A, C, W, and Y (3). However, serogroup B has presented unique challenges in vaccine development due to the similarity between the NmB capsular polysaccharide and a human neural cell adhesion molecule, which creates the risk of generating an autoimmune reaction (4, 5). The reverse vaccinology approach, starting from the genomic information determined for the bacterium, has enabled the identification of noncapsular protein surface antigens that could help prevent invasive meningococcal disease (IMD) caused by any capsular serogroup (6, 7). These included neisserial adhesin A (NadA), neisserial heparin-binding antigen (NHBA), and factor H-binding protein (fHbp), which, in combination with New Zealand strain outer membrane vesicles (NZ OMV) harboring the PorA P1.4 antigen, constitute the N. meningitidis serogroup B 4-component (MenB-4C) vaccine (Bexsero; GSK). MenB-4C was the first broad-coverage MenB vaccine based on recombinant proteins (8) and is the only one approved (in January 2013) for use in individuals 2 months of age and above by the European Medicines Agency (8). In January 2015, the U.S. Food and Drug Administration approved this vaccine for use in persons 10 to 25 years of age. Subsequently, the U.S. Advisory Committee on Immunization Practices (ACIP) recommended the use of MenB vaccines for all persons >10 years of age in certain high-risk groups and suggested its use for adolescents 16 to 23 years of age (9, 10).
Efficacy studies to demonstrate the benefit of meningococcal vaccines are not feasible due to the relatively low incidence of the disease. Efficacy of glycoconjugate vaccines against serogroups A, C, Y, and W was estimated using the serum bactericidal antibody assay with rabbit (rSBA) or human complement (hSBA) (11, 12) accepted as a surrogate of protection in the clinical evaluation of meningococcal vaccines (13, 14).
Extrapolating this approach to NmB poses significant challenges. The meningococcal genome’s plasticity facilitates adaptation of surface structures to changing environments through a variety of genetic mechanisms (15–17) and generates a high variability of sequence and level of surface expression for protein antigens, affecting strain susceptibility to vaccine-induced antibodies. To predict the effectiveness of a MenB vaccine, a large panel of bacterial isolates representative of meningococcal disease in the United States would need to be tested in hSBA versus a significant number of subjects, thus requiring large volumes of serum and human complement. This would be a challenging undertaking, especially in infant populations.
To overcome these limitations, the meningococcal antigen typing system (MATS) was developed and standardized across public health laboratories worldwide (18, 19). The MATS combines a sandwich enzyme-linked immunosorbent assay (ELISA) to measure the immunologic cross-reactivity and quantity of antigen expression for three MenB-4C protein antigens (fHbp, NadA, and NHBA) with genetic typing of the PorA variable 2 (VR2) region to determine the immune recognition potential for the OMV component. MATS typing for each antigen correlates with killing in the hSBA and predicts the susceptibility of NmB strains to killing in the hSBA, i.e., the strain coverage of the MenB-4C vaccine (18). MATS has been shown to be a conservative predictor of strain coverage by the MenB-4C vaccine in infants and adolescents (20). In addition, MATS coverage was shown to correlate with high rates of individual seroprotection (21).
MATS has been used to predict MenB-4C strain coverage in multiple countries, including Canada and countries in Europe (22–25), using cross-sectional panels of strain isolated from meningococcal disease cases in the respective countries during one or two epidemiological years. So far, however, the MenB-4C strain coverage has not been estimated in the United States; neither has the longitudinal stability of such predictions been examined.
In this study, we evaluated a longitudinal panel of 442 U.S. NmB disease isolates (collected in 2000 to 2008) that were selected by the Active Bacterial Core Surveillance (ABCs; CDC) laboratory that are representative of serogroup B meningococcal disease in the United States (26). MATS analysis was performed on each NmB isolate, and the results were correlated with antigen genotyping data to determine MenB-4C strain coverage in the United States and to analyze its longitudinal trends from 2000 to 2008.
RESULTS
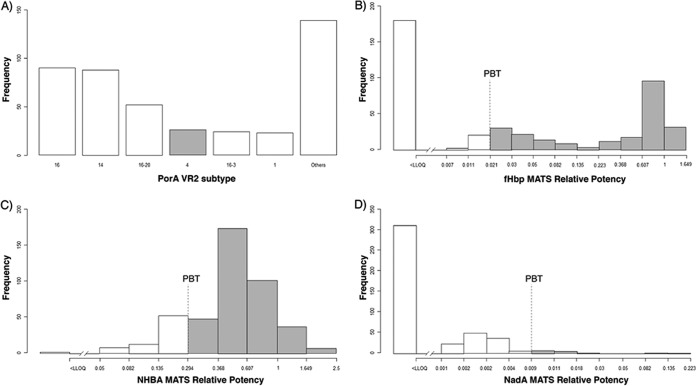
PorA VR2 subtype 4, covered by the OMV component of the MenB-4C vaccine (18), was identified in 23 strains (5.3%), while PorA subtypes 16 and 14 (each approximately 20%) predominated in 2000 to 2008 (Fig. 1A).
FIG 1 .
MenB-4C antigen frequency distribution in NmB U.S. isolates (n = 442). (A) Prevalence of PorA VR2 subtypes. Most of the NmB isolates belonged to the PorA 16 VR2 subtype (20.4%), followed by the PorA 14 VR2 subtype (19.9%). PorA VR2 subtype 4 is expressed in 5.9% of the isolates. (B) Frequency distribution of fHbp MATS relative potencies (RPs). Among the U.S. NmB isolates tested, 262/442 (59%) expressed fHbp above the MATS lower limit of quantitation (LLOQ) (gray bars). (C) Frequency distribution of NHBA MATS relative potencies (RPs) among the U.S. NmB isolates (n = 442). A total of 440 of the NmB isolates tested expressed NHBA above the MATS LLOQ (gray bars). (D) Frequency distribution of NadA MATS relative potencies (RPs). Among the U.S. NmB isolates tested, only 132 of 442 (30%) expressed NadA above the MATS LLOQ (gray bars).
Among the NmB U.S. isolates tested, the specific genotype of fHbp protein included in MenB-4C, variant 1 and peptide 1, was identified in 144 of 442 strains (33%). Among the fHbp variant 1 results, the most common peptides were peptides 1, 13, and 4 (33%, 7.2%, and 5.0%, respectively) (see Table 2). The most common factor H-binding protein variant 2 peptides were peptides 19, 24, and 16 (11%, 7.2%, and 5.0%, respectively); all other variant 2 and 3 peptides were identified in less than 5% of isolates (see Table 2). MATS detected quantifiable fHbp expression in 262/442 (59%) isolates (Fig. 1B). The NmB isolates with nonquantifiable MATS relative potencies (RP) for fHbp harbored fHbp variants 2 and 3, which were not included in the MenB-4C vaccine. NHBA expression was detected in 440 (95%) NmB isolates with a quantifiable MATS RP. NHBA peptides 5, 20, 10, 29, 3, and 21 were predominant in the U.S. NmB isolates (see Table 3). The specific genotype of NHBA included in the MenB-4C vaccine is peptide 2. This specific genotype was identified in only 42 of 442 (9.5%) isolates, but MATS demonstrated the immune reactivity of anti-NHBA antibodies with the majority (n = 440) of test isolates, irrespective of the NHBA genotype (Fig. 1C).
TABLE 2 .
Distribution of most frequent factor H-binding protein variant and peptide identities in the panel of 442 NmB strains
| Variant | fHbp peptide IDa |
Frequency (n = 442) |
% of total | Predicted coverage (95% CI) by fHbp |
Predicted coverage (95% CI) by all antigens |
|---|---|---|---|---|---|
| 1 | 1b | 144 | 33 | 99 (99–99) | 99 (99–99) |
| 13 | 25 | 5.7 | 24 (0–76) | 68 (52–92) | |
| 4 | 22 | 5.0 | 100 (91–100) | 100 (100–100) | |
| 14 | 17 | 3.8 | 94 (47–100) | 100 (94–100) | |
| 110 | 8 | 1.8 | 100 (100–100) | 100 (100–100) | |
| 12 | 6 | 1.4 | 100 (17–100) | 100 (67–100) | |
| Other IDs | 42 | 9.5 | 81 (57–86) | 100 (74–100) | |
| Total for variant 1 | 264 | 60 | 89 (77–95) | 97 (89–99) | |
| 2 | 19 | 48 | 11 | 0 | 94 (56–98) |
| 24 | 32 | 7.2 | 0 | 91 (41–100) | |
| 16 | 22 | 5.0 | 0 | 82 (27–86) | |
| 21 | 13 | 2.9 | 0 | 92 (62–100) | |
| 25 | 11 | 2.5 | 9.1 (0–9.1) | 55 (36–64) | |
| 3 | 31 | 7 | 1.6 | 0 | 86 (0–86) |
| 30 | 5 | 1.1 | 0 | 80 (60–100) | |
| 76 | 5 | 1.1 | 0 | 100 (100–100) | |
| 2/3 | Other IDs | 35 | 7.9 | 0 | 66 (43–83) |
| Total for variants 2 and 3 | 178 | 40 | 0.5 (0–0.5) | 83 (46–91) |
ID, identification number in PubMLST Neisseria sequence typing database. fHbp, factor H-binding protein.
Subvariant included in MenB-4C multicomponent vaccine.
TABLE 3 .
Distribution of the most frequent neisserial heparin-binding antigen peptides in the panel of 442 strains
| NHBA peptide IDa | Frequency (n = 442) | % of total | Predicted coverage (95% CI) by NHBA | Predicted coverage (95% CI) by all antigens |
|---|---|---|---|---|
| 5 | 127 | 29% | 87% (31%–100%) | 100% (98%–100%) |
| 20 | 43 | 9.7% | 98% (53%–100%) | 98% (63%–100%) |
| 2b | 42 | 9.5% | 100% (93%–100%) | 100% (100%–100%) |
| 10 | 29 | 6.6% | 97% (55%–100%) | 97% (55%–100%) |
| 29 | 26 | 5.9% | 92% (31%–100%) | 96% (42%–100%) |
| 3 | 26 | 5.9% | 100% (88%–100%) | 100% (96%–100%) |
| 21 | 25 | 5.7% | 84% (40%–92%) | 88% (40%–96%) |
| 1 | 18 | 4.1% | 100% (100%–100%) | 100% (100%–100%) |
| Other (n = 47) | 106 | 24% | 50% (21%–71%) | 69% (41%–84%) |
ID, identification number in PubMLST Neisseria sequence typing database.
Peptide included in the MenB-4C multicomponent vaccine.
The nadA gene was harbored by 170/442 (39%) strains, and 78% of these isolates (132/170) expressed NadA with quantifiable MATS RP (Fig. 1D).
Figure 2 summarizes the results of the MenB-4C strain coverage analysis performed. Results indicated that 91% (95% confidence interval [CI95], 72% to 96%) of the NmB U.S. isolates are predicted to be covered by the MenB-4C vaccine. While 5.7% (25/442) of the isolates had three antigens covered (RP > positive bacterial threshold [PBT] and/or PorA = P1.4), 42% (184/442) and 44% (194/442) had two antigens and one antigen covered, respectively (Fig. 2A). NHBA alone or in combination with another antigen(s) had RP values that were greater than the PBT values determined for 83% (CI95, 45% to 93%) of the NmB isolates followed by fHbp with 53% (CI95, 46% to 57%). The proportions of MATS-predicted coverage for PorA and NadA were 5.9% and 2.5% (CI95, 1.1% to 5.2%), respectively (Fig. 2B). In the case of NadA, the low contribution may have been due to the growth conditions in use for MATS testing, which repress nadA expression (27, 28). Having four antigens and two possible states (MATS positive or negative) each, 24=16 antigen combinations or MATS phenotypes can be observed. As shown in Fig. 2C, only 5 of the 16 phenotypes were observed with a frequency of >1%, accounting overall for 96% of the panel. The most frequent MATS phenotype was the double-positive phenotype for fHbp and NHBA antigens, followed by NHBA positive, negative for all antigens, fHbp positive, and triple positive for fHbp, NHBA, and PorA.
FIG 2 .

MenB-4C vaccine strain coverage potential among the U.S. NmB isolates (n = 442). (A) Among the U.S. NmB isolates tested, 194/442 (44%) expressed one antigen (1Ags), 184/442 (42%) two antigens (2Ags), and 25/442 (5.7%) three antigens (3Ags) at levels over the PBT thresholds for the respective antigens. Overall, MATS estimates 91% coverage of the 4CMenB vaccine among U.S. NmB isolates. 0Ags, no antigens. (B) Contribution of individual antigens to 4CMenB coverage of the U.S. NmB isolates (n = 442). NHBA conferred maximum coverage potentials, with 83% of NHBA-positive strains exhibiting PBT values over the threshold, followed by fHbp (53%), PorA (5.9%), and NadA (2.5%). (C) Frequency of MATS phenotypes among the U.S. NmB isolates. The most frequent combination was represented by MATS double positives for fHbp and NHBA antigens (39%) followed by MATS single positives for NHBA antigen (36%). The MATS-negative phenotype accounted for 9%.
Data corresponding to predicted strain coverage by year of strain isolation in the United States from 2000 to 2008 are shown in Fig. 3 (see also Table S1 in the supplemental material). Point estimates of predicted strain coverage were similar across different years, and the 95% CIs largely overlapped. Lower levels of strain coverage were seen in the isolates collected in 2006 and 2008 (88%) and 2000 (89%); higher levels of strain coverage were seen in 2001 (97%) and 2004 (95%). A 2-sided chi-square test was used to test the differences across years in the predicted MATS coverage and in the upper and lower limit of the 95% CIs. No statistical significant difference across years was observed (P value = 0.567, P value = 0.457, and P value = 0.654, respectively). A linear regression model was used to test the strain coverage stability over time, and no statistically significant trend was observed either (P value = 0.222; Table S2), indicating high and consistently stable strain coverage in the United States over almost a decade.
FIG 3 .

MATS coverage of NmB strains isolated in the United States during 2000 to 2008 by at least 1 antigen (95% CI) by year. Comparisons of point estimates of predicted strain coverage across groups did not identify any statistically significant difference (P = 0.567).
MATS coverage of NmB strains isolated in the United States in 2000 to 2008 by at least 1 antigen (95% CI) classified by year of strain isolation. Download TABLE S1, PDF file, 0.1 MB (54.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Linear regression results for the analysis of temporal trends. Download TABLE S2, PDF file, 0.05 MB (51.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data corresponding to the predicted strain coverage classified by clonal complex (CC) are shown in Table 1. Two major CCs accounting for more than half of the circulating strains (CC32 and CC41/44) had predicted strain coverage of 99% (CI95, 98% to 100% and 73% to 100%, respectively). The two other CCs with frequencies of >5%, CC162 and CC35, also had high levels of strain coverage (91% and 85%, respectively) but with broader confidence intervals (CI95, 53% to 100% and 29% to 92%, respectively), suggesting antigenic variability. Minor CCs had high strain coverage (between 50% and 89%), too.
TABLE 1 .
Distribution of clonal complexes in the panel of 442 NmB strains
| Clonal complex | Frequency (n = 442) |
% of total | Predicted % coverage (95% CI) by all antigens |
|---|---|---|---|
| CC32 | 162 | 37 | 99 (98–100) |
| CC41/44 | 123 | 28 | 99 (73–100) |
| CC162 | 43 | 9.7 | 91 (53–100) |
| CC35 | 24 | 5.4 | 85 (29–92) |
| CC269 | 19 | 4.3 | 68 (32–89) |
| CC60 | 9 | 2.0 | 89 (67–100) |
| CC213 | 6 | 1.4 | 50 (33–50) |
| Unassigneda | 25 | 5.7 | 64 (32–76) |
| Other (n = 14) | 31 | 7.0 | 68 (55–84) |
Unassigned, the sequence type (ST) of strains in this collection has been identified, but the ST belongs to no clonal complex (CC).
Tables 2 and 3 report predicted strain coverage by genotype of the fHbp and NHBA antigens, respectively. Within fHbp variant 1 (60% of the panel), fHbp covered >90% of all major peptides corresponding to identification numbers (IDs) in the PubMLST Neisseria sequence typing database with the exception of ID 13 (fHbp coverage, 24% [CI95, 0% to 76%]; 6% of the isolates) and 81% of the minor IDs, for an overall 89% coverage by fHbp. Within fHbp variants 2 and 3 (40% of the panel), fHbp provided almost no coverage, and yet each antigenic variant was significantly (50% to 100%) covered by other MenB-4C antigens, for an overall 83% coverage by non-fHbp MenB-4C antigens. Within the eight major NHBA peptide IDs (IDs 5, 20, 2, 10, 29, 3, 21, and 1, accounting for three-quarters of the circulating strains), NHBA provided consistently high (84% to 100%) coverage. Coverage by NHBA for minor peptide IDs was also high (69% [CI95, 41% to 84%]).
Due to the low number of isolates with levels of NadA MATS relative potency above the positive bacterial threshold (PBT; see Materials and Methods), the relationship between strain coverage and antigen genotype was not investigated for this antigen.
DISCUSSION
Molecular epidemiology is a component of bacterial surveillance programs that is important for understanding the incidence and diversity of the Neisseria meningitidis isolates collected in the United States. In order to predict the possible vaccine coverage potential, it is critical to assess the expression of vaccine antigens in the representative N. meningitidis isolates. A major limitation of the molecular approach to determine the potential for strain coverage by a vaccine is that it has not been possible thus far to obtain information about the expression levels of the protein encoded in the bacteria from genetic data. Expression of the protein is necessary if the bacteria are to be targeted by protective antibodies directed against the antigen of interest. Studies to characterize the geographic and temporal distribution of the vaccine antigens among the members of the N. meningitidis population in the United States are important to understand the disease dynamics and strategies for deployment of MenB vaccines. Considering the importance of antigen diversity and level of expression in predicting MenB-4C vaccine coverage, MATS was used to assess the expression and cross-reactivity to vaccine-induced antibodies of MenB-4C antigens among the isolates selected in this study as representative of NmB meningococcal disease in the United States.
MATS predicted MenB-4C vaccine coverage of over 90% of circulating NmB strains in the United States. For the first time globally, strain coverage was estimated for a significant period of time and was found to be consistent over a 9-year time period (2000 to 2008). Among the MenB-4C vaccine antigens, NHBA alone was shown to contribute to strain coverage in over 80% of the isolates in a consistent manner across antigen genotypes. In contrast, fHbp coverage potential (53%) was restricted to the variant 1 antigen genotypes, as the NmB isolates harboring fHbp variants 2 and 3 were MATS negative for this antigen. Nonetheless, 83% of the fHbp variant 2/3 strains were covered by other MenB-4C antigens, suggesting the importance of the multiple vaccine components. MATS represents a conservative predictor of strain coverage in relation to hSBA as it does not take into account synergy of combinations of antigens or the contribution of other components present in the outer membrane vesicles (20, 29). Also, MATS does not factor in the interactions between antigens present at levels below the PBT. This is particularly relevant for MenB-4C vaccine as it contains multiple components that may provide a greater synergistic effect even when the individual components are present at levels below the bactericidal threshold (30).
Genetic diversity among the endemic NmB isolates is well documented (24). This diversity is also seen in virulence genes that are vaccine targets. Possible changes in the expression profiles of these genes in response to genotypic variations cannot be ruled out. A change in the virulence factor’s structure might impact its immune reactivity, jeopardizing the positive outcome of vaccination. Evidence for this can be drawn from the MATS data obtained in this study with reference to fHbp, whose immune reactivity was restricted by the variant class of this gene that corresponds to vaccine antigen. Only those strains expressing variant 1 and its subclasses had expressed immune susceptibility to the vaccine. In contrast, NHBA, despite the molecular variations in the gene that encode the target peptides, was found to show immune reactivity to antibodies induced by the major and minor peptide IDs.
Microbes are under constant evolutionary pressure posed by several factors, including vaccines. Given the possibility of genotypic changes in a particular virulence gene in response to vaccine pressure, an ideal vaccine should have inherent fail-safe mechanisms. One such strategy is to create a multicomponent vaccine such as MenB-4C. The presence of multiple components in MenB-4C vaccine implies that coverage may be maintained for strains that express multiple antigens in the event of mutation or loss of expression of a single antigen. It was previously noted that the bactericidal activity is increased for strains expressing two or more of the vaccine antigens at levels higher than the positive bactericidal threshold (18).
Although this study was limited to the evaluation of NmB strains circulating up to 2009, the stability of the strain coverage prediction over the 9-year period investigated suggests that no major differences should be expected in the ensuing years. Also, all genetic lineages significantly represented in the country were covered at levels of >85%, suggesting that temporal oscillations in the frequency of clonal complexes would not affect significantly the MenB-4C strain coverage. Postimplementation surveillance capable of monitoring both genetic (antigens or full-genome sequencing) and phenotypic (MATS) variations will be the key to quick adaptation of the public health strategy to the potential response of the pathogen to vaccination (31), especially when joined with dynamic modeling techniques that quantify in nearly real time the effectiveness of immunization campaigns (32). Also, adopting the same tools and standards for bacterial typing implementation worldwide, earlier epidemiological evidence from one country could help in preventive adjustments of the implementation strategy in other areas.
In September 2015, the United Kingdom was the first country to introduce MenB-4C into the national infant immunization program, offering the vaccine to all infants born after 1 July 2015, with a 2-plus-1 schedule at 2, 4, and 12 months and with two small catch-up cohorts for infants born between 1 May and 30 June (3, 4, and 12 months and 4 and 12 months). A detailed multifaceted plan is in place for enhanced meningococcal disease surveillance in England that will provide invaluable data on the usefulness of MATS for monitoring vaccine impact, characterizing meningococci causing meningococcal disease in both vaccinated and unvaccinated cohorts and the impact on meningococcal disease in infants. The reduced infant schedule implemented from September 2015 in the United Kingdom national immunization program provided the first evidence of effectiveness of 83% for a two-dose vaccine against all NmB disease, equivalent to a vaccine effectiveness of 94% against the most highly predicted NmB disease-preventable strains (33). This vaccine effectiveness measured in the field exceeds significantly the MATS coverage predictions for the United Kingdom (67% in 2014/2015, representing the last year prior to mass vaccination). Considering the higher (91%) MATS strain coverage predictions in the United States, the effectiveness of MenB-4C vaccination in the United States could be potentially superior to the UK results, particularly considering the burden of disease in the infant population for which a direct link was established between MATS strain coverage and pooled hSBA titers and individual seroprotection (21). Recently, the FDA approved the use of MenB-4C before licensure to control several outbreaks in U.S. universities (34). The FDA subsequently licensed MenB-4C for the age group of 10 to 25 years, and the ACIP recommended its use for all persons of age 10 and older in high-risk groups (9). No cases of meningococcal disease caused by N. meningitidis serogroup B have been reported among vaccinated students, although the numbers were too low to allow statistically powered determinations of efficacy (34). A recent immunogenicity study conducted among U.S. college students during a NmB outbreak indicated that 87% to 100% of vaccinated students showed immunoreactivity to reference NmB strains compared to modest (66.1%) reactivity to the NmB outbreak strain (35). This difference in the levels of immune reactivity among the MenB-4C seropositives reconfirms our incomplete understanding of strain susceptibility to vaccine-induced bactericidal activity and the tendency of hSBA to underestimate the vaccine efficacy (36). Additional data regarding the breadth and duration of protection provided by meningococcal B vaccine will be key in the decision-making process pertaining to vaccine dosage and schedule.
This report has provided a detailed breakdown of molecular characterization and MATS results for U.S. invasive NmB strains, estimating a high level of strain coverage for the MenB-4C vaccine in the United States. Overall, the data provide a useful baseline for monitoring MenB-4C. However, due to anticipated changes in the distribution of clonal complexes and antigen genotypes over time, continuous detailed surveillance and monitoring of the antigen expression of circulating strains will be needed.
MATERIALS AND METHODS
Strain selection and classification.
A representative panel of 442 U.S. NmB isolates collected by the Active Bacterial Core surveillance (ABCs; CDC) in 2000 to 2008 were tested in MATS. Among these U.S. isolates, representation of fHbp 1.1 was weighted to account for higher rates of serogroup B and overall meningococcal disease occurring in Oregon. Multilocus sequence types, clonal complexes, and antigen genotypes for these isolates had previously been determined (26). The U.S. isolates (n = 442) tested in this study included the major clonal complexes of endemic U.S. NmB strains. Clonal complexes CC32 (37%), CC41/44 (28%), and CC162 (9.7%) accounted for the majority of the strains (Table 1).
Genotypic classification of antigens.
The classification of fHbp followed the scheme available on the PubMLST Neisseria sequence typing database (http://pubmlst.org/neisseria/), which separates peptide subvariants into three major variant classifications: variants 1, 2, and 3. Unique NHBA peptides and nadA variants were numbered as previously described (37, 38). PorA subtyping had been previously performed by PCR amplification and sequencing of the VR2 region of the gene (26). PorA variants had been assigned therein according to variable region 2 (VR2) sequences on the PubMLST Neisseria sequence typing database.
Meningococcal antigen typing system (MATS) ELISA.
The MATS ELISA methodology (18), reagents, reference strains, and recombinant antigens were supplied by Novartis Vaccines and Diagnostics, Siena, Italy (now part of the GSK group of companies). Briefly, bacteria were cultured overnight on chocolate agar (BD Biosciences, San Jose, CA) and were suspended in Mueller-Hinton (MH) broth (BD Biosciences, San Jose, CA) to an optical density at 600 nm (OD600) of 0.4. Empigen BB detergent (Sigma, St. Louis, MO) was added to the bacterial suspension to reach a final dilution of 1:11, and this suspension was incubated at 45°C for 1 h for bacterial inactivation. Twofold serial dilutions of bacterial extract in MH broth with Empigen were carried out in ELISA plates (Costar, Corning, NY) that had been precoated with rabbit polyclonal antibodies against fHbp, NHBA, or NadA. The plates were incubated for 1 h at 37°C and then washed with phosphate-buffered saline (PBS)–0.05% Tween. The plates were incubated for 1 h at 37°C with biotinylated antigen-specific rabbit polyclonal antibody to detect the primary antibody-bound antigens. After being washed, the plates were developed with streptavidin-horseradish peroxidase (HRP) (Jackson ImmunoResearch, West Grove, PA) and o-phenylenediamine dihydrochloride (OPD; Sigma). The reaction was stopped with 4N H2SO4 and read at 492 nm. A reference strain for each antigen (H44/76 for fHbp, NGH38 for NHBA, and 5/99 for NadA) was also included in the respective microtiter plates. Results were analyzed with StatLIA software (Brendan Technologies, Carlsbad, CA). The relative potency (RP) for each unknown strain was calculated by comparing the fit of the five-parameter logistic regression curves to 2-fold serial dilutions of extracts from the reference strain and the unknown strain. The reference strain for each antigen was assigned an arbitrary value of 100.
Estimation of NmB strain coverage.
In a previous study, it was shown that the presence of at least one antigen with a relative potency (RP) value greater than the positive bactericidal threshold (PBT; 0.021 for fHbp, 0.294 for NHBA, and 0.009 for NadA) or the presence of PorA P1.4 correlated with bactericidal activity in the hSBA by pooled sera taken after immunization (18). Therefore, the MATS data could be used to predict strain coverage by the multicomponent MenB-4C vaccine. Strains that did not meet these criteria were deemed not covered. In this study, the predicted strain panel coverage was defined as the proportion of strains with a MATS relative potency value greater than the positive bactericidal threshold value for one or more antigens and/or with PorA P1.4. It should be noted that the PBT was set using pooled sera from infants and that strain coverage predictions do not account for variations in the individual seroresponses.
Statistical analysis.
As described in the MATS interlaboratory standardization study (19), empirical estimates of the 95% CIs for the positive bactericidal thresholds were derived with a log-normal approximation based on the overall ranges of assay reproducibility (0.014 to 0.031 for fHbp, 0.169 to 0.511 for NHBA, and 0.004 to 0.019 for NadA). These values were used to define the 95% CIs of the strain coverage data. No CI was calculated for coverage by the PorA antigen, as the typing was performed genotypically (VR2 = P1.4).
All statistical evaluations were performed using R statistical software (http://www.r-project.org) version 2.13.1.
ACKNOWLEDGMENTS
Amanda C. Cohn (CDC) provided necessary guidance for the selection of NmB strains with Oregon strain weighting. Members of the Bacterial Meningitis Reference Laboratory of the CDC provided the NmB strains used in this study.
Material and support for this study were provided by GlaxoSmithKline Biologicals SA. GlaxoSmithKline Biologicals SA was involved in analysis and interpretation of data. GlaxoSmithKline Biologicals SA was provided the opportunity to review a preliminary version of the manuscript for factual accuracy, but we are solely responsible for final content and interpretation. We received no financial support or other form of compensation related to the development of the manuscript.
M.S., G.B., L.S., and D.M. are currently employed by the GSK group of companies. G.R., E.K., and P.S. have performed research on behalf of the Centers for Disease Control and Prevention (Atlanta, GA, USA). G.C. was employed at the CDC at the time of the study.
G.R., M.S., D.M., and G.C. participated in the conception and design of the study. E.K., P.S., and G.B. participated in the acquisition of data. All of us participated in the analysis and interpretation of the data and in the drafting and revision of the manuscript.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Khatami A, Pollard AJ. 2010. The epidemiology of meningococcal disease and the impact of vaccines. Expert Rev Vaccines 9:285–298. doi: 10.1586/erv.10.3. [DOI] [PubMed] [Google Scholar]
- 2.Giuliani MM, Adu-Bobie J, Comanducci M, Aricò B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, Cartocci E, Ciucchi L, Di Marcello F, Ferlicca F, Galli B, Luzzi E, Masignani V, Serruto D, Veggi D, Contorni M, Morandi M, Bartalesi A, Cinotti V, Mannucci D, Titta F, Ovidi E, Welsch JA, Granoff D, Rappuoli R, Pizza M. 2006. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snape MD, Pollard AJ. 2005. Meningococcal polysaccharide-protein conjugate vaccines. Lancet Infect Dis 5:21–30. doi: 10.1016/S1473-3099(04)01251-4. [DOI] [PubMed] [Google Scholar]
- 4.Finne J, Bitter-Suermann D, Goridis C, Finne U. 1987. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J Immunol 138:4402–4407. [PubMed] [Google Scholar]
- 5.Stein DM, Robbins J, Miller MA, Lin FY, Schneerson R. 2006. Are antibodies to the capsular polysaccharide of Neisseria meningitidis group B and Escherichia coli K1 associated with immunopathology? Vaccine 24:221–228. doi: 10.1016/j.vaccine.2005.07.084. [DOI] [PubMed] [Google Scholar]
- 6.Pizza M, Scarlato V, Masignani V, Giuliani MM, Aricò B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 7.Rappuoli R. 2000. Reverse vaccinology. Curr Opin Microbiol 3:445–450. doi: 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 8.Vernikos G, Medini D. 2014. Bexsero® chronicle. Pathog Glob Health 108:305–316. doi: 10.1179/2047773214Y.0000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folaranmi T, Rubin L, Martin SW, Patel M, MacNeil JR, Centers for Disease Control (CDC) . 2015. Use of serogroup B meningococcal vaccines in persons aged ≥10 years at increased risk for serogroup B meningococcal disease: recommendations of the Advisory Committee on Immunization Practices, 2015. MMWR Morb Mortal Wkly Rep 64:608–612. [PMC free article] [PubMed] [Google Scholar]
- 10.MacNeil JR, Rubin L, Folaranmi T, Ortega-Sanchez IR, Patel M, Martin SW. 2015. Use of serogroup B meningococcal vaccines in adolescents and young adults: recommendations of the Advisory Committee on Immunization Practices, 2015. MMWR Morb Mortal Wkly Rep 64:1171–1176. doi: 10.15585/mmwr.mm6441a3. [DOI] [PubMed] [Google Scholar]
- 11.Borrow R, Carlone GM. 2001. Serogroup B and C serum bactericidal assays. Methods Mol Med 66:289–304. doi: 10.1385/1-59259-148-5:289. [DOI] [PubMed] [Google Scholar]
- 12.Maslanka SE, Gheesling LL, Libutti DE, Donaldson KB, Harakeh HS, Dykes JK, Arhin FF, Devi SJ, Frasch CE, Huang JC, Kriz-Kuzemenska P, Lemmon RD, Lorange M, Peeters CC, Quataert S, Tai JY, Carlone GM. 1997. Standardization and a multilaboratory comparison of Neisseria meningitidis serogroup A and C serum bactericidal assays. The Multilaboratory Study Group. Clin Diagn Lab Immunol 4:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldschneider I, Gotschlich EC, Artenstein MS. 1969. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med 129:1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin L, Gilbert PB, Corey L, McElrath MJ, Self SG. 2007. A framework for assessing immunological correlates of protection in vaccine trials. J Infect Dis 196:1304–1312. doi: 10.1086/522428. [DOI] [PubMed] [Google Scholar]
- 15.Budroni S, Siena E, Dunning Hotopp JC, Seib KL, Serruto D, Nofroni C, Comanducci M, Riley DR, Daugherty SC, Angiuoli SV, Covacci A, Pizza M, Rappuoli R, Moxon ER, Tettelin H, Medini D. 2011. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A 108:4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Siena E, D’Aurizio R, Riley D, Tettelin H, Guidotti S, Torricelli G, Moxon ER, Medini D. 2016. In-silico prediction and deep-DNA sequencing validation indicate phase variation in 115 Neisseria meningitidis genes. BMC Genomics 17:843. doi: 10.1186/s12864-016-3185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch C, Moxon ER, Stella M, Comanducci M, Bambini S, Muzzi A, Andrews W, Chen J, Santos G, Santini L, Boucher P, Serruto D, Pizza M, Rappuoli R, Giuliani MM. 2010. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci U S A 107:19490–19495. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plikaytis BD, Stella M, Boccadifuoco G, DeTora LM, Agnusdei M, Santini L, Brunelli B, Orlandi L, Simmini I, Giuliani M, Ledroit M, Hong E, Taha MK, Ellie K, Rajam G, Carlone GM, Claus H, Vogel U, Borrow R, Findlow J, Gilchrist S, Stefanelli P, Fazio C, Carannante A, Oksnes J, Fritzsønn E, Klem AM, Caugant DA, Abad R, Vázquez JA, Rappuoli R, Pizza M, Donnelly JJ, Medini D. 2012. Interlaboratory standardization of the sandwich enzyme-linked immunosorbent assay designed for MATS, a rapid, reproducible method for estimating the strain coverage of investigational vaccines. Clin Vaccine Immunol 19:1609–1617. doi: 10.1128/CVI.00202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frosi G, Biolchi A, Lo Sapio M, Rigat F, Gilchrist S, Lucidarme J, Findlow J, Borrow R, Pizza M, Giuliani MM, Medini D. 2013. Bactericidal antibody against a representative epidemiological meningococcal serogroup B panel confirms that MATS underestimates 4CMenB vaccine strain coverage. Vaccine 31:4968–4974. doi: 10.1016/j.vaccine.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Budroni S, Kleinschmidt A, Boucher P, Medini D. 2016. Pooled-sera hSBA titres predict individual seroprotection in infants and toddlers vaccinated with 4CMenB. Vaccine 34:2579–2584. doi: 10.1016/j.vaccine.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Bettinger JA, Scheifele DW, Halperin SA, Vaudry W, Findlow J, Borrow R, Medini D, Tsang R; Members of the Canadian Immunization Monitoring Program, Active (IMPACT) . 2013. Diversity of Canadian meningococcal serogroup B isolates and estimated coverage by an investigational meningococcal serogroup B vaccine (4CMenB). Vaccine 32:124–130. doi: 10.1016/j.vaccine.2013.03.063. [DOI] [PubMed] [Google Scholar]
- 23.Křížová P, Musílek M, Vacková Z, Kozáková J, Claus H, Vogel U, Medini D. 2014. Predicted strain coverage of a new protein-based meningococcal vaccine in the Czech Republic. Epidemiol Mikrobiol Imunol 63:103–106. [PubMed] [Google Scholar]
- 24.Vogel U, Taha MK, Vazquez JA, Findlow J, Claus H, Stefanelli P, Caugant DA, Kriz P, Abad R, Bambini S, Carannante A, Deghmane AE, Fazio C, Frosch M, Frosi G, Gilchrist S, Giuliani MM, Hong E, Ledroit M, Lovaglio PG, Lucidarme J, Musilek M, Muzzi A, Oksnes J, Rigat F, Orlandi L, Stella M, Thompson D, Pizza M, Rappuoli R, Serruto D, Comanducci M, Boccadifuoco G, Donnelly JJ, Medini D, Borrow R. 2013. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: a qualitative and quantitative assessment. Lancet Infect Dis 13:416–425. doi: 10.1016/S1473-3099(13)70006-9. [DOI] [PubMed] [Google Scholar]
- 25.Medini D, Stella M, Wassil J. 2015. MATS: global coverage estimates for 4CMenB, a novel multicomponent meningococcal B vaccine. Vaccine 33:2629–2636. doi: 10.1016/j.vaccine.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Cohn A, Comanducci M, Andrew L, Zhao X, MacNeil JR, Schmink S, Muzzi A, Bambini S, Rappuoli R, Pizza M, Murphy E, Hoiseth SK, Jansen KU, Anderson AS, Harrison LH, Clark TA, Messonnier NE, Mayer LW. 2011. Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine 29:4739–4744. doi: 10.1016/j.vaccine.2011.04.092. [DOI] [PubMed] [Google Scholar]
- 27.Fagnocchi L, Pigozzi E, Scarlato V, Delany I. 2012. In the NadR regulon, adhesins and diverse meningococcal functions are regulated in response to signals in human saliva. J Bacteriol 194:460–474. doi: 10.1128/JB.06161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metruccio MM, Pigozzi E, Roncarati D, Berlanda Scorza F, Norais N, Hill SA, Scarlato V, Delany I. 2009. A novel phase variation mechanism in the meningococcus driven by a ligand-responsive repressor and differential spacing of distal promoter elements. PLoS Pathog 5:e1000710. doi: 10.1371/journal.ppat.1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weynants VE, Feron CM, Goraj KK, Bos MP, Denoël PA, Verlant VG, Tommassen J, Peak IR, Judd RC, Jennings MP, Poolman JT. 2007. Additive and synergistic bactericidal activity of antibodies directed against minor outer membrane proteins of Neisseria meningitidis. Infect Immun 75:5434–5442. doi: 10.1128/IAI.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vu DM, Wong TT, Granoff DM. 2011. Cooperative serum bactericidal activity between human antibodies to meningococcal factor H binding protein and neisserial heparin binding antigen. Vaccine 29:1968–1973. doi: 10.1016/j.vaccine.2010.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snape MD, Medini D, Halperin SA, DeTora L, Drori J, Moxon ER. 2012. The challenge of post-implementation surveillance for novel meningococcal vaccines. Vaccine 30(Suppl 2):B67–B72. doi: 10.1016/j.vaccine.2011.12.126. [DOI] [PubMed] [Google Scholar]
- 32.Argante L, Tizzoni M, Medini D. 2016. Fast and accurate dynamic estimation of field effectiveness of meningococcal vaccines. BMC Med 14:98. doi: 10.1186/s12916-016-0642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parikh SR, Andrews NJ, Beebeejaun K, Campbell H, Ribeiro S, Ward C, White JM, Borrow R, Ramsay ME, Ladhani SN. 2016. Effectiveness and impact of a reduced infant schedule of 4CMenB vaccine against group B meningococcal disease in England: a national observational cohort study. Lancet 388:2775–2782. doi: 10.1016/S0140-6736(16)31921-3. [DOI] [PubMed] [Google Scholar]
- 34.McNamara LA, Shumate AM, Johnsen P, MacNeil JR, Patel M, Bhavsar T, Cohn AC, Dinitz-Sklar J, Duffy J, Finnie J, Garon D, Hary R, Hu F, Kamiya H, Kim HJ, Kolligian J, Neglia J, Oakley J, Wagner J, Wagner K, Wang X, Yu Y, Montana B, Tan C, Izzo R, Clark TA. 2015. First use of a serogroup B meningococcal vaccine in the US in response to a university outbreak. Pediatrics 135:798–804. doi: 10.1542/peds.2014-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basta NE, Mahmoud AAF, Wolfson J, Ploss A, Heller BL, Hanna S, Johnsen P, Izzo R, Grenfell BT, Findlow J, Bai X, Borrow R. 2016. Immunogenicity of a meningococcal B vaccine during a university outbreak. N Engl J Med 375:220–228. doi: 10.1056/NEJMoa1514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granoff DM. 2016. Meningococcal B vaccine during a university outbreak. N Engl J Med 375:1594. doi: 10.1056/NEJMc1610666. [DOI] [PubMed] [Google Scholar]
- 37.Bambini S, Muzzi A, Olcen P, Rappuoli R, Pizza M, Comanducci M. 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27:2794–2803. doi: 10.1016/j.vaccine.2009.02.098. [DOI] [PubMed] [Google Scholar]
- 38.Lucidarme J, Comanducci M, Findlow J, Gray SJ, Kaczmarski EB, Guiver M, Vallely PJ, Oster P, Pizza M, Bambini S, Muzzi A, Borrow R. 2010. Characterization of fHbp, nhba (gna2132), nadA, porA, and sequence type in group B meningococcal case isolates collected in England and Wales during January 2008 and potential coverage of an investigational group B meningococcal vaccine. Clin Vaccine Immunol 17:919–929. doi: 10.1128/CVI.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MATS coverage of NmB strains isolated in the United States in 2000 to 2008 by at least 1 antigen (95% CI) classified by year of strain isolation. Download TABLE S1, PDF file, 0.1 MB (54.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Linear regression results for the analysis of temporal trends. Download TABLE S2, PDF file, 0.05 MB (51.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.