FIG 2 .
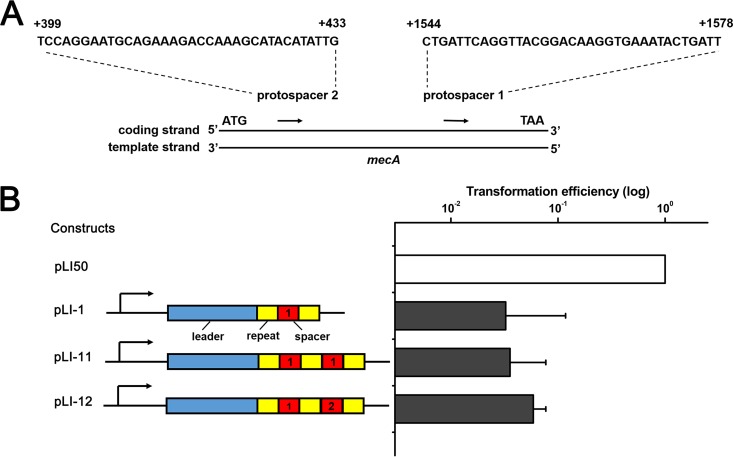
One chromosome-targeting spacer is sufficient for CRISPR targeting. (A) Schematic of two sequence regions selected as an artificial CRISPR array-targeting site. Sequences of the coding strand from 1544 to 1578 nt and from 399 to 433 nt relative to the start codon (ATG) of mecA constituted protospacer 1 and protospacer 2, respectively. (B) mecA-targeting constructs pLI-1 (one spacer), pLI-11 (two identical spacers), and pLI-12 (two different spacers) displayed similar toxicity. The transformation efficiency of the empty plasmid pLI50 (no spacer) was set at 100%. Transformations were performed three times, and average relative transformation efficiencies plus standard deviations (error bars) are shown in the graph.