Figure 6. Superposition of KAI2 and QDO shows structural conservation and functional diversity.
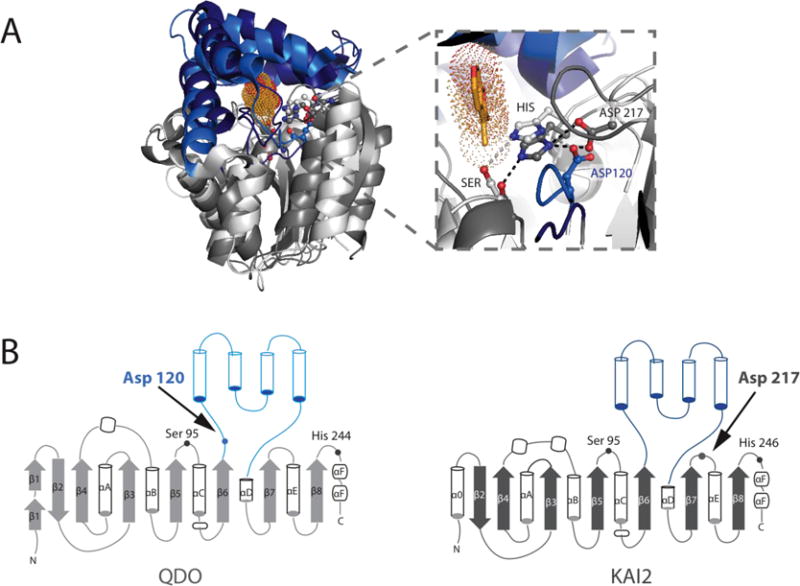
A) Sequence-independent structural superposition of A. thaliana KAI2 (PDB: 3JKM) and Pseudomonas putida 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (QDO, PDB: 3IBT), with core domains shown in dark and light grey, and the lid domains shown in dark and light blue, respectively. Karrikin is in bright orange sticks with dots in the KAI2 active site. Zoom in of the active site highlights that despite the distance in primary sequence, the two enzymes have their catalytic triad Aspartates in largely the same location, enabling efficient H-bonding with their respective catalytic Histidines. B) Topology diagrams showing various differences in the core domains of both enzymes and the differences in Catalytic Aspartate placement within the structure.
