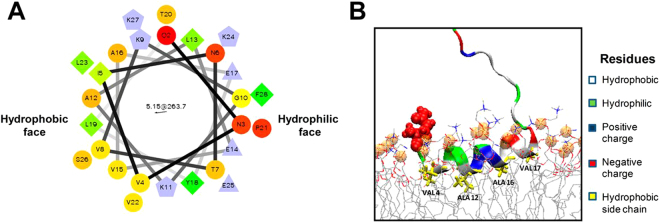
Figure 1.
Predicted structure of ATG3 N-terminal amphipathic α-helix. (A) Helical wheel representation of the predicted N-terminal amphipathic helix (amino acids 2–27) of human ATG3 (wheel generated in Helical Wheel Projections). (B) The predicted insertion of ATG3 N-terminal α-helix into membranes. Hydrophobic (Leu, Ala, Val), negatively-charged (Glu) and positively-charged (Lys) residues are coloured yellow, red and blue respectively (courtesy of Dr. Martin B. Ulmschneider, Johns Hopkins University, Baltimore, MD). Peptide binding onto a lipid bilayer was studied by unbiased atomic detail molecular dynamics simulations following a protocol described in detail elsewhere59. In brief, the peptide was initially placed in bulk water in a fully extended conformation and allowed to freely fold and bind to the membrane. No biasing potentials were applied. The peptide was found to bind irreversibly and fold onto the interface within one microsecond.