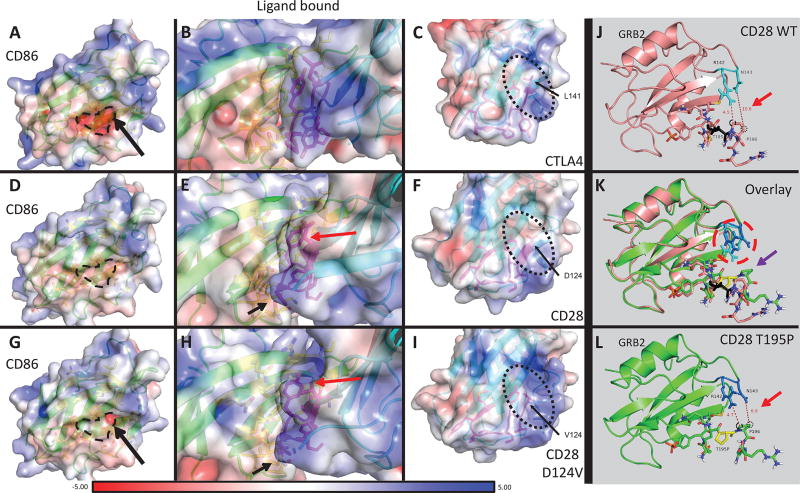
Figure 3. Protein modeling of CD28 WT and D124V interacting with CD86, and CD28 WT and T195P interacting with GRB2.
A–I: PyMOL models of CD86 bound to receptor CTLA4, CD28 WT, or CD28 D124V with APBS-generated electrostatic surfaces. A–C: CD86-CTLA4 interaction. The residue corresponding to D124 in CD28 (L141) is indicated in C and the protein surface in the vicinity of this residue has an overall positive charge (also apparent in B). The negatively charged binding surface of CD86 can be seen in A (dashed line and surrounding area). D–F: CD86-CD28 WT interaction. The surface around D124 is more neutral compared to CTLA4. G–I: CD86-CD28 D124V interaction. Replacement of the negatively charged aspartate residue leads to a more positively charged surface, similar to CTLA4. The orientation of several residues is changed (Y118 – red arrow, P121 – black arrow) between CD28 WT and CD28 D124V in these models. J-L: PyMOL models of the CD28 cytoplasmic tail with or without the T195P mutant binding adaptor protein GRB2. J, L: CD28 WT (J) and T195P (L) cytoplasmic tail binding GRB2 (J, salmon; L, chartreuse). T195 is colored black (J); T195P is colored yellow (L). Note the distances between CD28 P196 carbons Cβ (black dotted circle) or Cδ (black dashed circle) and the closest non-hydrogen atoms on GRB2 (red arrows). CD28 P196 Cβ moves from 10.6Å away from GRB2 N143 in the CD28 WT to 6.9Å in CD28 T195P. K: Overlay of CD28 WT versus T195P mutant binding of GRB2. There is a significant change in orientation of several CD28 residues: proximal SH3-domain P196 (purple arrow) rotates and approaches GRB2 N143 in the CD28 T195P mutant. GRB2 R142 and N143 have a strikingly different rotation in the CD28 T195P mutant compared to WT (red dashed circle).