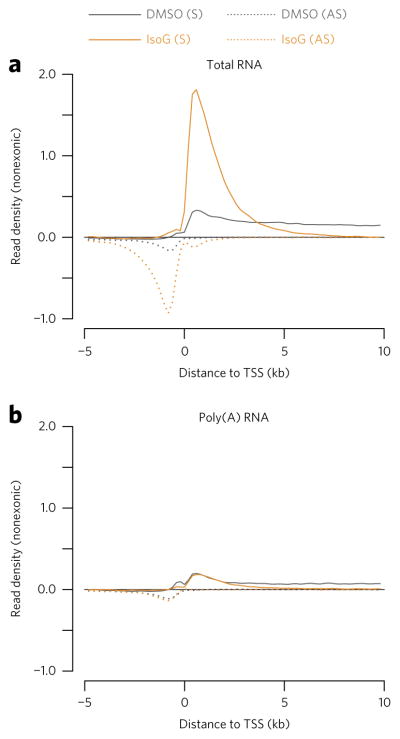
Figure 1. Strand-specific total RNA-seq reveals global accumulation of nonpolyadenylated promoter-proximal RNA transcripts upon isoginkgetin (IsoG) treatment.
(a,b) Metaplots showing mean nonexonic read density across annotated promoter regions, aligned at transcription start sites (TSSs) and normalized to ERCC spike-in controls. (a) Strand-specific total RNA-seq in which ribosomal RNAs were depleted from total RNA by hybridization. (b) mRNA-seq in which poly(A)+ RNAs were purified from total RNA by oligo-dT hybridization. The traces represent an average of n = 2 replicates of 30 μM IsoG treatment of HeLa cells for 6 h. In these and subsequent TSS metaplots, only genes longer than the maximum x-axis value (here 10 kb) are included. S, sense; AS, antisense.