FIGURE 2.
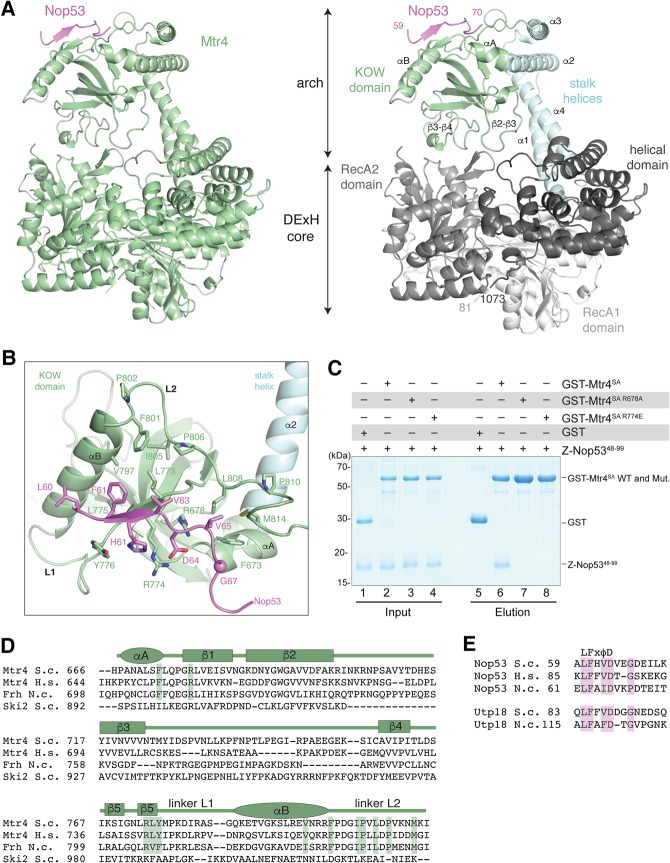
Crystal structure of S. cerevisiae Mtr4-ΔN bound to the Nop53 AIM. (A) On the left is the overall structure of Mtr4-ΔN (green) and the Nop53 AIM motif (pink). On the right is a more detailed snapshot where Mtr4 is in the same orientation but colored by domains (as in the schematics in Fig. 1A). The RecA1, RecA2, and helical domains of the DExH core are colored from lighter to darker shades of gray. The stalk helices and KOW domain of the arch are in cyan and green, respectively. Secondary structure elements discussed in the text are highlighted. (B) Zoom-in view of the interactions between Nop53 and the Mtr4 KOW domain. The domains are colored as in Figure 2A, right panel, and viewed after a 90° rotation around a horizontal axis with respect to the view in Figure 2A. Residues discussed in the text are highlighted and labeled. (C) Protein coprecipitations by GST pull-down assays. GST-tagged yeast Mtr4 short arch (GST-Mtr4SA), and mutants were purified and mixed with purified Z-tagged Nop5348-99. Pull-down assays were carried out using GSH-Sepharose beads in a buffer containing 150 mM NaCl. The Coomassie-stained 16% SDS–PAGE gels show the input (lanes 1–4) and the pulled-down protein precipitates (lanes 5–8). (D) Sequence alignment of the Mtr4 KOW domain from S. cerevisiae (S.c.), H. sapiens (H.s.), and the N. crassa (N.c.) ortholog FRH. The alignment includes the related cytoplasmic helicase Ski2 from S. cerevisiae. Secondary structure elements are shown above the sequence alignment with α-helices indicated by an ellipse and β-strands by a rectangle. (E) Sequence alignment of the arch-interacting motifs (AIM) of Nop53 and Utp18 from S. cerevisiae (S.c.), H. sapiens (H.s.), and the N. crassa (N.c.). Note: H.s. UTP18 does not contain an AIM and is therefore not shown.