Summary
The LM609 antibody specifically recognizes αVβ3 integrin and inhibits angiogenesis, bone resorption, and viral infections in an arginine-glycine-aspartate independent manner. LM609 entered phase II clinical trials for the treatment of several cancers and was also used for αVβ3-targeted radio-immunotherapy. To elucidate the mechanisms of recognition and inhibition of αVβ3 integrin, we solved the structure of the LM609 antigen-binding fragment by X-ray crystallography and determined its binding affinity for αVβ3. Using single-particle electron microscopy we show that LM609 binds at the interface between the β-propeller domain of the αV chain and the βI domain of the β3 chain, near the RGD-binding site, of all observed integrin conformational states. Integrating this data with fluorescence size-exclusion chromatography, we demonstrate that LM609 sterically hinders access of large ligands to the RGD binding pocket, without obstructing it. This work provides a structural framework to expedite future efforts utilizing LM609 as a diagnostic or therapeutic tool.
Graphical Abstract
The LM609 antibody specifically recognizes αVβ3 integrin and has been employed in numerous clinical and academic research applications. In the manuscript, The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human αVβ3 Integrin via Steric Hindrance, Borst AJ et al. shed light on the mode-of-action of a widely used integrin-targeting antibody.
Introduction
Integrins are heterodimeric cell-surface receptors mediating bidirectional signaling across the plasma membrane. They are assembled from non-covalently associated α and β subunits and are involved in the physiological processes of cellular adhesion, migration, growth, immunity, and differentiation (Shimaoka and Springer, 2003). Regulation of integrin activity occurs through large-scale conformational changes following ligand-binding to either the cytoplasmic or extracellular domains of the integrin heterodimer. Integrin activation entails transition from a bent, low-activity conformation, to an extended state with a significantly increased affinity for extracellular ligands (Campbell and Humphries, 2011; Ye et al., 2010). Among all integrins, αVβ3 has been highly studied for its roles in arthritis, thrombosis, and inflammation, as well as its localized expression to neovasculature and aggressive cancerous tumors (Xiong et al., 2001; Xiong et al., 2002). In addition, many viruses have been reported to use αVβ3 integrin as a receptor or co-receptor for cellular attachment and subsequent entry (Stewart and Nemerow, 2007; Veesler et al., 2014).
Given its biomedical significance, αVβ3 has emerged as a promising therapeutic target for several human diseases (Danhier et al., 2012; Gutheil et al., 2000). Recent efforts in drug development have targeted the αVβ3 Arg-Gly-Asp (RGD) binding pocket formed at the interface between the β-propeller of the αV subunit and the βI domain of the β3 subunit within the integrin headpiece (Xiong et al., 2002). αVβ3 recognition of RGD-containing ligands, such as vitronectin, fibronectin, and fibrinogen, results in rapid integrin clustering leading to platelet activation, increased bone resorption, elevated immune response, and neovascularization (Koo et al., 2002). Synthetic RGD-peptide-based strategies have yielded molecules with high affinity for the integrin RGD-binding pocket, but generally lacking integrin specificity. Since the RGD motif is recognized by nearly half of all 24 known integrin receptor types, non-specific pharmacologic recognition of numerous integrins can lead to unintended side effects.
Therapeutic antibodies provide a promising alternative, and account for most integrin-targeting drugs currently under clinical evaluation. In addition to their high specificity, therapeutic antibodies are advantageous due to their low immunogenicity and longer half-life in circulating blood relative to many small molecules and peptides (Millard et al., 2011). However, many antibodies currently used in pre-clinical and clinical trials independently target either the integrin α or β subunit, leading to recognition of any integrin heterodimer containing the target subunit (Frelinger et al., 1991). Of notable exception is LM609, a monoclonal murine antibody specific for integrin αVβ3 (Wu et al., 1998). Preclinical studies have shown that LM609 inhibits angiogenesis, bone resorption, and viral pathogenesis both in vitro and in vivo (Garrigues et al., 2008; Gavrilovskaya et al., 1999; Gavrilovskaya et al., 1998; Gramoun et al., 2007; Posey et al., 2001; Triantafilou et al., 2000). A fully humanized variant of LM609, MED-523 (Vitaxin), entered Phase I clinical trials for the treatment of cancers. Results of these trials reported no significant toxicity at any dose level (Gutheil et al., 2000). Phase II clinical trials for the treatment of colorectal cancer showed disease stabilization in 7 out of 14 patients with one patient experiencing partial regression (Gutheil et al., 2000). The second generation of Vitaxin, known as Etaracizumab (Abegrin), demonstrated increased binding affinity to αVβ3 with a significant reduction in brain tumor size following αVβ3-targeted radio-immunotherapy (Veeravagu et al., 2008). Further studies demonstrated Abegrin can act as an effective agent for αVβ3-positive tumor-imaging (Liu et al., 2011). Abegrin has since entered Phase II clinical trials for the treatment of lymphoma, melanoma, renal, small intestine, and colorectal cancers, but the results of many of these trials currently remain unreported.
We sought to elucidate the molecular determinants associated with LM609 function and its mechanism of action. To this end, we determined the structure of the LM609 antigen-binding fragment (Fab) using X-ray crystallography and characterized its binding kinetics to human αVβ3 integrin. Single particle electron microscopy allowed for structural characterization of the complex formed between the Fab LM609 and the αVβ3 integrin ectodomain. Our results indicate LM609 binds with high-affinity to the apex of the integrin headpiece near but without obstructing the RGD-binding site, as confirmed by fluorescence size-exclusion chromatography. Comparison with a crystal structure of the αVβ3/fibronectin complex suggests that LM609 prevents binding of RGD-containing ligands via steric hindrance which explains the reported inhibition of angiogenesis and bone resorption mediated by this antibody in preclinical and clinical studies.
Results
Crystal Structure of LM609
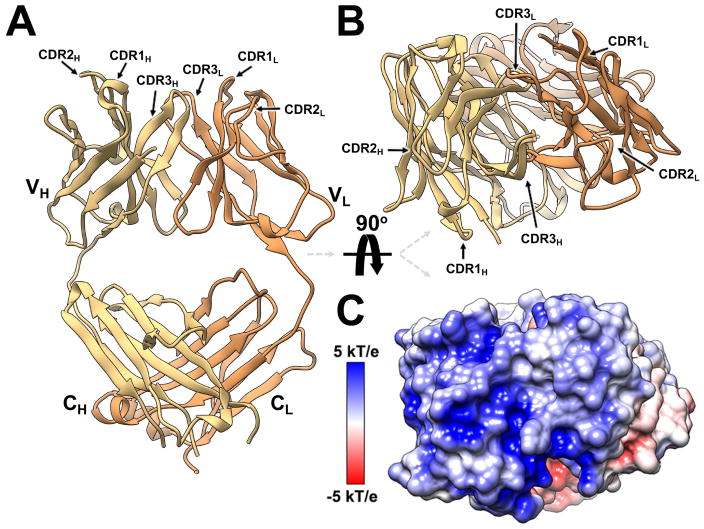
The structure of Fab LM609 was determined at 2.3 Å resolution using X-ray crystallography (Fig. 1A–B, Table S1). Calculation of the LM609 electrostatic properties revealed that the surface formed at the apex of the Fab, containing the complementarity-determining regions (CDRs), is characterized by a marked positive potential (Fig. 1C). This is explained by the presence of numerous basic residues directly contributed by the CDRs and by the flanking immunoglobulin framework regions. Furthermore, the amino acid composition of the LM609 CDRs follows previously reported trends with numerous aromatic, glycine and serine residues (Fellouse et al., 2007; Fellouse et al., 2004; Sidhu and Kossiakoff, 2007). The Fab elbow angle, defined as the angle between the pseudo-2-fold axes relating the two variable and the two constant domains, was determined to be 148.6°. This structure provides a framework to rationalize a previously reported directed evolution study aimed at enhancing the affinity of Vitaxin for αVβ3 integrin (Wu et al., 1998) and for future structure-guided design studies using this antibody as a therapeutic platform.
Figure 1. Crystal structure of the Fab LM609.
(A–B) Ribbon diagrams showing two orthogonal views of the Fab LM609 colored yellow and orange for the heavy and light chains, respectively. The complementarity-determining regions (CDRs) are indicated. VH: heavy chain variable domain; CH: heavy-chain constant domain; VL: light chain variable domain; CL: light-chain constant domain. (C) Surface representation of LM609 colored by electrostatic potential showing the marked positive nature of the Fab apex. The orientation is identical to the one in panel B.
The Fab LM609 Binds with Nanomolar Affinity to αVβ3 Integrin
We used surface plasmon resonance to characterize the affinity of Fab LM609 to the ectodomain of human αVβ3 integrin. Immobilization of αVβ3 integrin was achieved through amine coupling with measurements performed using Fab concentrations ranging between 7.8 and 500 nM (Figure 2A). Fab LM609 kinetic association (kon) and dissociation constants (koff) for αVβ3 were determined to be 1.5 ± 0.26 × 105 M−1s−1 and 3.4 ± 0.002 × 10−3 s−1, respectively, yielding an equilibrium dissociation constant (KD) of 2.3 ± 0.6 × 10−8 M. αVβ3 binding kinetics reported here are in agreement with the previously published kinetics for the humanized therapeutic variant of Fab LM609, Vitaxin (Wu et al., 1998). These results confirm grafting of murine CDRs onto the human antibody framework had minimal impact on the binding properties of this molecule.
Figure 2. Characterization of the Fab LM609/αVβ3 integrin complex.
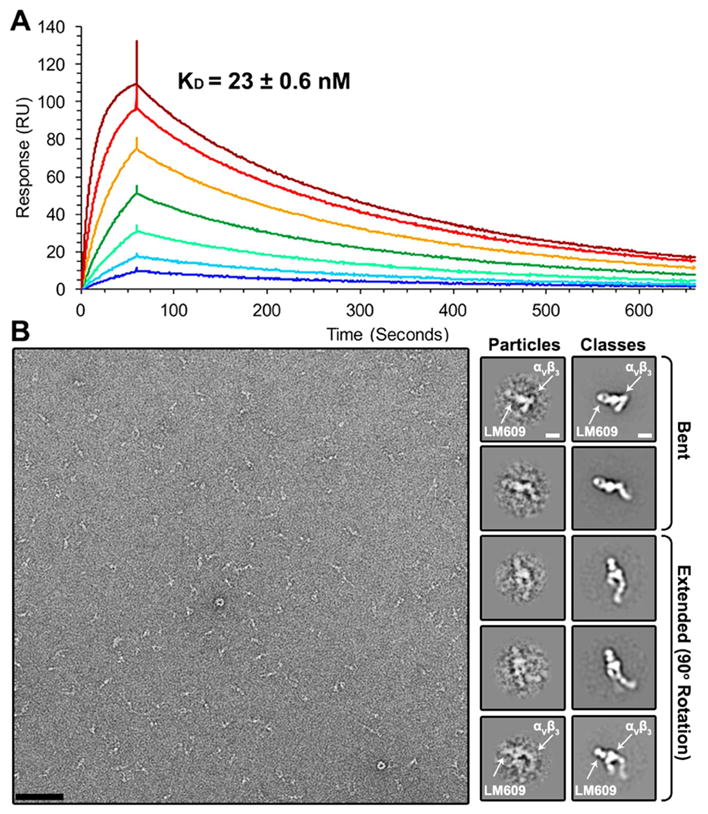
(A) Kinetic analysis of LM609 binding to αVβ3 using surface plasmon resonance. Each curve on the sensorgram corresponds to a different concentration of LM609 (analyte): 7.8 nM (dark blue); 15.6 nM (light blue); 31.2 nM (light green); 62.5 nM (dark green); 125 nM (orange); 250 nM (red); 500 nM (dark red). (B) (Left) Raw electron micrograph of negatively stained Fab LM609/αVβ3 integrin complex. (Right) Raw particles (low-pass filtered to 20 Å resolution) and resulting 2D class averages of various conformations of the integrin/LM609 complex. Scale bars: micrograph: 652 Å; class averages: 60 Å.
LM609 Specifically Binds to the αVβ3 Integrin Headpiece Region
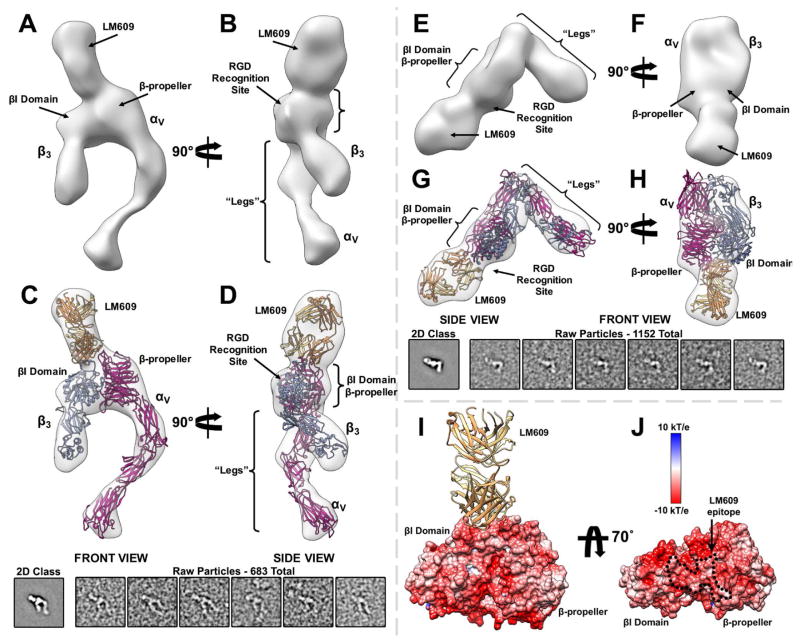
Multiple structural studies have demonstrated the αVβ3 heterodimer can assume a bent, inactive conformation with the integrin “leg” domains wrapped around the headpiece. In addition, several different extended conformations representative of different stages of integrin activation have also been reported (Takagi et al., 2002; Xiao et al., 2004; Xiong et al., 2001; Xiong et al., 2002). Imaging of the complex formed between human αVβ3 integrin and the Fab LM609 using negative staining electron microscopy revealed LM609 binds to all observed, bent or extended, αVβ3 conformational states (Fig. 2B). This result suggests that complex formation with Fab LM609 does not require the αVβ3 ectodomain to be in a defined conformational state.
An iterative computational strategy was used to obtain homogeneous 2D class averages corresponding to various integrin conformational snapshots (Lyumkis et al., 2014; Veesler et al., 2014). In all resolved conformations, LM609 interacts with the αVβ3 headpiece domain (Fig. 2B). The orientation of the Fab relative to αVβ3 was determined from the 2D class averages by identification of the typical Fab top and side views bound to the closed and open integrin conformations, respectively (Wu et al., 2012). Analysis of 2D class averages suggests the LM609 epitope is at the interface between the β-propeller domain of the αV chain and the βI domain of the β3 chain (Fig. 2B); both domains could be distinguished due to the typical triangular shape of the headpiece apex region. In contrast, attempted complex formation between Fab LM609 and αIIbβ3 and visualization using negative staining EM yielded no detectable complexes (Fig. S1). This result corroborates the early observation that LM609 specifically binds to αVβ3 but not to αIIbβ3 (Wu et al., 1998), in conflict with a recent report suggesting LM609 may bind solely to the β3 integrin subunit (Kamata et al., 2013).
We used the random-conical tilt (RCT) approach to compute 3D reconstructions from each independent 2D class average of the integrin conformational ensemble (Radermacher, 1988). The resulting density maps (Fig. 3A–B & E–F, Supplementary Movie 1, and Fig. S2 A & C) are in excellent agreement with the corresponding class averages with observed αVβ3 conformers readily assigned to known integrin functional states (Chen et al., 2010; Takagi et al., 2002; Zhu et al., 2008). We obtained pseudo-atomic models of the αVβ3/LM609 complex by fitting the Fab LM609 crystal structure reported here along with a previously determined αVβ3 integrin crystal structure (Xiong et al., 2001; Xiong et al., 2002) into the 3D reconstructions using Rosetta CM (Song et al., 2013) (Fig. 3C–D & G–H and Fig. S2 A). These maps provide enough features to fit the LM609 Fab and determine its approximate position and orientation relative to the integrin headpiece. The 3D models support the proposed location of the LM609 epitope on the αVβ3 integrin headpiece apex at the interface between the β-propeller and βI domains. However, the 2-fold pseudo-symmetry of LM609 prevents unequivocal placement of the Fab relative to the integrin apex due to the limited resolution of the RCT reconstructions. Electrostatic surface potential calculations of αVβ3 integrin show that the headpiece apex, which includes the LM609 epitope, features a predominantly negatively charged surface (Fig. 3I–J). It is likely that the electrostatic complementarity of the paratope and epitope contributes to the high binding-affinity of LM609 for the αVβ3 integrin apex.
Figure 3. Structure of the Fab LM609/αVβ3 integrin complex determined by single particle EM.
(A–B) Two orthogonal views of a random conical tilt 3D reconstruction featuring an αVβ3 integrin activated state. (C–D) Corresponding views related to (A–B) showing the fit of the pseudo-atomic model obtained with Rosetta (ribbon) into the reconstruction (transparent surface). The inset included below depicts the associated 2D class average and a few raw particles (low-pass filtered to 20 Å resolution) used for the RCT reconstruction of the integrin/LM609 extended-state. (E–F) Two orthogonal views of a random conical tilt 3D reconstruction featuring an αVβ3 integrin bent state. (G–H) Corresponding views related to (E–F) showing the fit of the pseudo-atomic model obtained with Rosetta (ribbon) into the reconstruction (transparent surface). The inset included below depicts the associated 2D class average and a few raw particles (low-pass filtered to 20 Å resolution) used for the RCT reconstruction of the integrin/LM609 bent-state. (I) Ribbon diagram of the Fab LM609 bound to the integrin headpiece. Only the β-propeller of the αV subunit and the βI domain of the β3 subunit are shown in surface representation colored by electrostatic potential. (J) Corresponding view related to (I) rotated by 70°. The approximate epitope of the LM609 Fab is indicated with black dashed lines and features a pronounced negative electrostatic potential. LM609 is represented as yellow and orange ribbons corresponding to the heavy and light chains, respectively. αVβ3 is represented as fuchsia (αV) and light blue (β3) ribbons in panels (C–D) and (G–H).
The LM609 Epitope does not Overlap with the RGD Ligand Binding Pocket
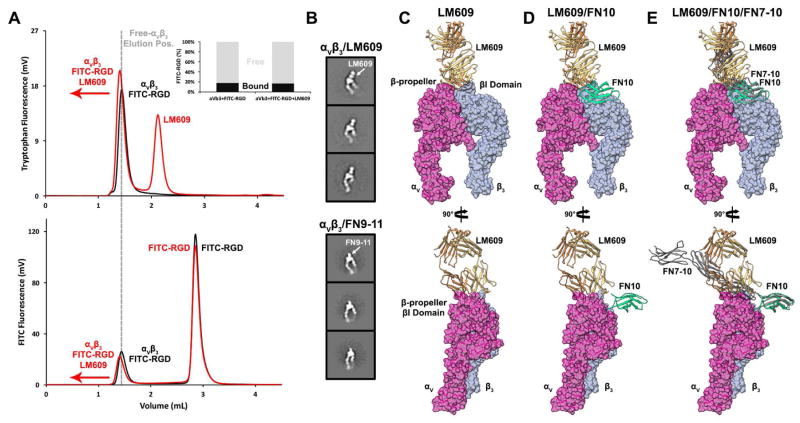
Previous reports have shown that LM609 prevents binding of extracellular ligands to the RGD-binding pocket of αVβ3 integrin (Charo et al., 1990; Cheresh, 1987). Since the LM609 Fab does not appear to overlap with the RGD binding pocket in any of the integrin conformations observed, we set out to elucidate the mechanism associated with competitive binding. We used fluorescence size-exclusion chromatography (Hattori et al., 2012; Kawate and Gouaux, 2006) to determine if αVβ3 can bind to a fluorescein isothiocyanate-tagged RGD peptide, FITC-GRGDSPK (FITC-RGD), in the absence or presence of bound LM609 Fab (Fig. 4A). As expected, FITC-RGD could bind to integrin αVβ3 in the absence of LM609. FITC-RGD could also bind to αVβ3, with the same magnitude, following pre-incubation of αVβ3 with a saturating amount of Fab LM609. The earlier elution volume observed for the αVβ3/FITC-RGD/LM609 peak relative to the αVβ3/FITC-RGD peak combined with tryptophan and FITC-RGD fluorescence data confirmed both ligands could simultaneously bind to αVβ3, ruling out competitive binding to the same epitope. We next showed that pre-incubation of αVβ3 with a cyclic RGD peptide prevented FITC-RGD binding, demonstrating competition would occur between LM609 and FITC-RGD if they were competing for the same epitope (Fig. S3). Although previous studies reported that LM609 inhibits RGD-mediated ligand binding (Charo et al., 1990; Cheresh, 1987), our data further clarifies that this antibody does not directly occlude the RGD-binding pocket. Instead, LM609 likely precludes binding of large RGD-ligands via steric hindrance.
Figure 4. Mechanism of αVβ3 integrin inhibition by the Fab LM609.
(A) FSEC profiles revealing FITC-RGD binds integrin αVβ3 in the presence of a saturating amount of LM609 Fab. (Top panel) Tryptophan fluorescence elution profile of α Vβ3/FITC-RGD in the absence (black) or presence (red) of LM609. (Bottom panel) FITC fluorescence elution profile of αVβ3/FITC-RGD in the absence (black) or presence (red) of LM609. The dotted grey line indicates the elution volume corresponding to αVβ3 in the absence of bound LM609. In both panels, binding of LM609 shifts integrin αVβ3 to earlier elution volumes. (Inset) Relative amounts of free (grey) and integrin αVβ3-bound (black) FITC-RGD peptide plotted in the absence and presence of LM609, demonstrating LM609 does not compete with small RGD-containing ligands for binding to αVβ3 integrin. (B) LM609 binds to αVβ3 integrin in the vicinity of the region mediating attachment of physiological ligands such as fibronectin. 2D class averages of extended αVβ3 integrin in the presence of LM609 (first column) or FN9-11 (second column). (C–E) Two orthogonal views of a surface representation of the αVβ3 headpiece with bound LM609 (B), with bound LM609 and the domain 10 of fibronectin type III (FN10) (C), and with bound LM609, FN10 and the domains 7–10 of fibronectin type III (FN7-10) (D). Pseudo-atomic models were derived by superimposing the β-propeller and β-I domains from the αVβ3/FN10 complex crystal structure (PDB 4MMX) onto the model presented in Figure 3. The crystal structure of FN7-10 (PDB 1FNF) was then superimposed onto the corresponding residues of FN10 to approximate its position and orientation relative to αVβ3 (B–D).
Discussion
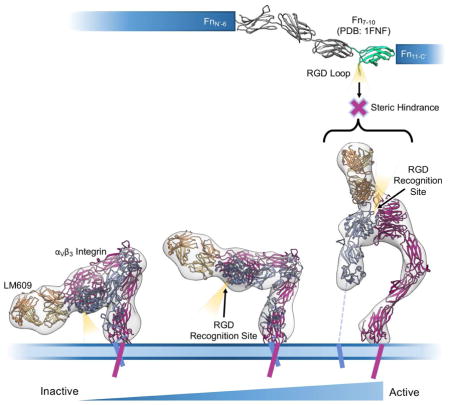
Our EM results demonstrate LM609 binds to the αVβ3 integrin headpiece apex in the vicinity of the region mediating attachment of physiological ligands such as vitronectin, fibrinogen, or fibronectin (Fig. 4B–E). Whereas numerous endogenous binders rely on an exposed RGD motif for interactions with integrin, no such motif is present in LM609. Although both RGD-independent (LM609) and RGD-dependent (e.g. fibronectin) binding involve contacts with the β-propeller of the αV subunit and the βI domain of the β3 subunit, distinct sets of interactions mediate attachment in each case (Fig. 4D). As a result, LM609 binding to αVβ3 does not directly occlude the RGD binding site. However, in vitro studies demonstrated LM609 inhibits fibronectin binding to αVβ3 (Charo et al., 1990). Superposition of the fibronectin type III repeats 7–10 structure (Leahy et al., 1996) (FN7-10, PDB 1FNF) onto the corresponding domain of the αVβ3 integrin/FN10 complex structure (Van Agthoven et al., 2014) (PDB 4MMX) reveals the mechanism underlying LM609-mediated inhibition of αVβ3 (Charo et al., 1990): binding of LM609 to the integrin headpiece would likely sterically hinder subsequent attachment of fibronectin due to the expected clashes with FN domains 8 and 9 (Fig. 4E and Fig. S4). Previous reports also showed LM609 can inhibit the binding of fibrinogen to αVβ3 integrin in vitro (Wu et al., 1998). The structural and biophysical data reported here suggest this is mediated through a mechanism of steric hindrance due to the comparably large sizes of fibrinogen and fibronectin (Mosesson, 2005), and the ability of the FITC-RGD peptide to bind to LM609-bound integrin αVβ3 (Fig. 4A). This inhibition mechanism allows rationalizing the properties of this antibody that has been shown to inhibit angiogenesis and bone resorption in vivo. These results also explain why LM609 inhibits infection of RGD-containing viruses such as Parechovirus and Kaposi’s Sarcoma-Associated Herpesvirus (Garrigues et al., 2008; Triantafilou et al., 2000). In our experiments, Fab LM609 could bind to all observed integrin conformational states, including the bent state. This argues that LM609 has the potential to interact with αVβ3 receptors prior to inside-out signal activation, which could be leveraged for future therapeutic strategies. Similarly to what has been proposed for the 17E6 antibody (Mahalingam et al., 2014), the full-length LM609 antibody might also interfere with integrin clustering or promote integrin internalization, therefore enhancing therapeutic effects relative to the monovalent Fab LM609.
Our results shed light on the previously cryptic binding mode of an antibody that has been extensively studied for academic, preclinical, and clinical research applications. Many current RGD-based anti-integrin drugs have low specificity and act primarily as agonists for all integrins recognizing the RGD motif (Van Agthoven et al., 2014). In contrast, the high specificity of LM609 for αVβ3 and the absence of selection for activated conformational states makes it a promising antagonistic candidate for future work directed towards αVβ3-positive cancerous tumors and as a competitive binder against RGD-containing viruses targeting integrin αVβ3 as a receptor for infection. Finally, the specificity of LM609 for αVβ3 raises the possibility of developing antibody-drug conjugates directing therapeutic compounds to αVβ3 integrin-expressing cell-types. We expect the data reported here will expedite efforts utilizing LM609 as a potential diagnostic or therapeutic tool for a variety of human diseases.
METHOD DETAILS
Protein Expression and Purification
The synthetic genes encoding human integrin αV and β3 subunits (NCBI accession codes NM_002210 and NM_000212), were optimized for expression in insect cells and purchased from Genscript. These genes were used as a template for PCR amplification of the regions encoding the ectodomains and comprising residues 31–987 and 27–718, for αV and β3 subunits, respectively (numbering corresponds to that of the immature proteins). Each amplified DNA fragment was cloned into pT350 vector (Krey et al., 2010), which contains BiP secretion signal peptide and the inducible metallothionein promoter, activated by divalent cations. Drosophila S2 cells were co-transfected with the plasmids carrying the αV and β3 ectodomain genes, and stably-transfected cell lines were established (Backovic and Krey, 2016). The protein expression was induced by 0.5mM CuSO4. Supernatants containing the αVβ3 ectodomain were collected 7 days after induction.
Recombinant Fab LM609 was also produced in Drosophila S2 cells, using the expression strategy developed for obtaining milligram quantities of recombinant Fabs (Backovic et al., 2010). Genes encoding the variable fragments of the light and heavy chains (VL and VH, respectively) of the mouse LM609 antibody (Rader et al., 1998), were optimized for expression in insect cells and cloned into a bicistronic vector, that already contained the mouse IgG1 constant regions CL1 and CH1. Protein secretion was driven by the BiP signal peptide, and the heavy chain was tagged with a double strep tag at the C-terminus, allowing affinity purification using streptactin resin. The Fab was further purified using a Superdex 75 size exclusion column run in a buffer containing 10mM Tris pH 8 and 50mM NaCl (Backovic et al., 2010).
To obtain integrin-Fab complexes, the supernatants of S2 cells expressing the integrin were mixed with purified Fabs, and run over streptactin column. The eluted material contained the mixture of integrin-Fab complexes and the excess, unbound Fab. The two forms were separated using size exclusion chromatography, and the fractions corresponding to the peak containing the complex were concentrated. Additional αVβ3 and αIIbβ3 ectodomain constructs were purchased from R&D Systems.
BL21DE3 cells containing the plasmid GST-FN9-11 pGEX6P1 were grown at 37°C until an OD600 of 0.7 was reached. Protein expression was induced overnight at 37°C using 1mM IPTG. Cells were harvested via centrifugation, resuspended in 20 mM Tris pH 7.4, 150 mM NaCl, 5 mM DTT, 0.1 mM PMSF and kept at 4°C for 30 minutes before sonication. The cell lysate was clarified via centrifugation at 14,000 RPM for 20 minutes at 4°C and the resulting supernatant was incubated overnight at 4°C with chronic nutation in the presence of 1.5 mL of GSH-Agarose resin. The resin was subsequently poured over a disposable column to remove unbound proteins. Elution was achieved with a buffer containing 50 mM Tris-HCl pH 8.0, 15 mM GSH after a 10-minute incubation period at 4°C. Fractions containing GST-FN9-11 were dialyzed into PBS at 4°C to remove excess GSH and DTT. Cleavage of GST was performed using PreScission protease for 1 hour at 4°C. Cleaved FN9-11 was further incubated for 2 hours at 4°C with 1.5 mL GSH-Agarose resin to allow removal of free GST and uncleaved GST-FN9-11. GSH-Agarose beads were centrifuged at 4,000 RPM for 20 minutes and purified FN9-11 was collected from the supernatant.
Crystal Structure of LM609
Recombinant LM609 Fab in 10mM Tris pH 8, 50mM NaCl was concentrated to 7.7 mg/ml and used for screening of crystallization conditions. Sitting drops were set up at 18°C by mixing equal volumes of protein a nd crystallization solutions. Crystals appeared in the condition containing 0.1M HEPES pH 7.5, 30% PEG 4,000 and 0.2M CaCl2, and were cryo-cooled by plunging into liquid nitrogen, after equilibration in the same buffer containing 20% glycerol as a cryoprotectant.
X-ray data were collected at the microfocus beamline PX2 at the French national synchrotron facility (SOLEIL), and processed using the XDS package (Kabsch, 2010). Molecular replacement was done using Phaser (McCoy et al., 2007) and Fab PDB code 2ADG as a search model(Zhou et al., 2005). An atomic model was built in Coot (Emsley et al., 2010) and refined using Buster (Blanc et al., 2004). The heavy and light chains of Fab LM609 contain 217 and 214 residues, respectively, and continuous electron density accounts for the majority of both chains. No density is observed for residues 133–135 of the heavy chain, and residues 1 and 214 of the light chain, which were omitted from the model.
Surface Plasmon Resonance
αVβ3 integrin was reconstituted from lyophilized powder at 0.5 mg/mL in 10 mM HEPES pH 8.0, 50 mM NaCl, 2 mM MnCl2, 0.05% P20 buffer. Amine coupling of 0.025mg/mL αVβ3 integrin was achieved in Sodium Acetate, pH 4.0 to a CM5 chip with a target immobilization of 1000 response units. The Fab LM609 was injected over the CM5 flow cells with a contact time of 60 seconds at a flow-rate of 60 μL/min. Fab LM609 concentrations used for kinetic analysis ranged between 7.8 nM and 500 nM. Fab LM609 dissociation from αVβ3 occurred over 600 seconds with a flow rate of 60 μL/min. Regeneration of αVβ3 was accomplished by pulsing 10 mM glycine, pH 1.5 for 4 seconds at a flow rate of 30 μL/min. Experiments were performed in triplicate for each concentration of Fab LM609 tested.
Negative-Stain EM Specimen Preparation
LM609/αVβ3 samples for random conical tilt experiments were negatively stained using 2% uranyl-formate on glow-discharged C-flat 2/0.5 grids overlaid with a thin layer of carbon. FN9-11/αVβ3, FN9-11/αIIbβ3, and LM609+αIIbβ3 samples were negatively stained on glow-discharged G400 Gilder grids in 10 mM Tris pH 8.0, 150 mM NaCl, 2mM MnCl2 using a five-fold molar excess of FN9-11 ligand and a 2.5 molar excess of Fab LM609. All integrin complex samples were deposited on carbon grids at a determined integrin concentration of 0.008 mg/mL.
EM Data Collection and Processing
All data were collected on an FEI Tecnai Spirit 120 kV electron microscope equipped with a Gatan Ultrascan 4000 CCD Camera. 509 image-pairs (0° and ± 55°) were collected using a random defocus range of −1.1 to −2.0 μm with a total exposure of 30 e−/Å2. The pixel size at the specimen level was 1.60 Å. Data collection was performed using Leginon (Suloway et al., 2005) with the majority of the data processing carried out in Appion (Lander et al., 2009). The parameters of the contrast transfer function (CTF) were estimated using CTFFIND3 (Mindell and Grigorieff, 2003). All particles were picked in a reference-free manner using DoG Picker (Voss et al., 2009). Correlation of particle tilt pairs was performed using Auto Tilt Picker (Voss et al., 2009). Particles were extracted with a binning factor of 2 with an initial box size of 288 pixels for the LM609/αVβ3 dataset for both the tilted and untilted particle datasets. FN9-11/integrin particles were extracted with a box size of 256 pixels. CTF-correction on the particles from the micrographs collected at 0° tilt was performed via phase-flipping with EMAN 1.9. The LM609/αVβ3 particle stack from the micrographs collected at 0° tilt was pre-processed in Relion 1.3 (Scheres, 2012) with an additional binning factor of 2 applied, resulting in a final box size of 72 pixels and a final pixel size of 6.4 Å. A binning factor of 2 was also applied to the FN9-11/integrin datasets, resulting in a final box size of 64 pixels at 6.4 Å/pix. 2D classification was carried out in Relion 1.3 (Scheres, 2012) on 93,992 particles from the micrographs collected at 0° tilt and clustered into 256 independent classes for the LM609 dataset. 27,325 selected particles were further sorted into 128 additional classes through an additional round of 2D classification. FN9-11/integrin complexes were sorted into 256 classes, with the best particles sorted into additional groups. 11 reference-free class averages representative of different integrin conformational states were centered with EMAN 2.04 (Tang et al., 2007) and uploaded to Appion as templates for reference-based alignment using SPIDER (Shaikh et al., 2008) on corresponding particle sub-stacks. Resulting alignments were further clustered using SPIDER Coran (Tang et al., 2007). Random conical tilt reconstructions were computed from the resulting clustered classes using the initial model pipeline built into Appion and particle images extracted from micrographs acquired at a 55° tilt angle. Final resolution estimates for all reported LM609/αVβ3 models varied between 35 and 55 Å using an FSC correlation of 0.5. Movie generation of the Fab LM609/αVβ3 conformational ensemble was performed on 3D models using the animation feature embedded within UCSF Chimera (Goddard et al., 2007). The 3D reconstructions were aligned based on the determined position, rotation, and motion of the leg domains during activation relative to the LM609/αVβ3 headpiece fiducial.
Fluorescence Size Exclusion Chromatography
The FITC-GRGDSPK peptide (96.7% purity, FITC-RGD) was purchased from GenScript and solubilized in ultrapure water. Integrins were prepared by dissolving lyophilized αVβ3 into 10 mM HEPES pH 8, 150 mM NaCl, 4mM MnCl2, 0.05% Tween 20. Samples for FSEC were prepared using a final αVβ3 integrin concentration of 100 nM in 10 mM Tris pH 8, 50 mM NaCl, 2 mM MnCl2. LM609 was added to corresponding integrin samples to a final concentration of 600 nM (6-fold molar excess) and allowed to incubate on ice for 30 minutes at 4°C. Subsequently, FITC-GRGDSPK was added at a final concentration of 600 nM (6-fold molar excess) to the appropriate aliquots of αVβ3 pre-incubated either in the presence or absence of 600 nM LM609 fab (1:1 FITC-RGD:LM609). Each sample was allowed to incubate for an additional 15 minutes, centrifuged at 16,200 × g for 10 minutes, and then transferred to a fresh 2 mL microfuge tube. Finally, all samples were injected onto a Superdex 200 Increase 5/150GL (GE Healthcare) pre-equilibrated with 10 mM Tris pH 8, 50 mM NaCl, 2 mM MnCl2 buffer while recording tryptophan fluorescence (λEx= 280 nm, λEm= 325 nm). Samples containing FITC-RGD peptide were reinjected and recorded using FITC fluorescence (λEx= 490 nm, λEm= 520 nm). Assessment of competition between FITC-RGD and cyclic RGD was carried out as a separate run. Cyclic RGD peptide was added to a final concentration of 600 nM (6-fold molar excess relative to αVβ3 integrin) and incubated on ice for 30 minutes at 4°C prior to addition of FITC-RGD peptide and subsequent FSEC analysis.
Model Generation
Pseudo-atomic models of the various conformations of the LM609/αVβ3 integrin complex were generated using Rosetta. Starting from the αVβ3 crystal structure (PDB ID: 3IJE) (Xiong et al., 2009) and the LM609 crystal structure, RosettaDock (Lyskov and Gray, 2008) augmented with an energy term assessing agreement to experimental density (DiMaio et al., 2009) was run. Due to the limited resolution of the RCT reconstructions, the 2-fold pseudo-symmetry of LM609 prevented unequivocal placement of the Fab relative to the integrin apex. However, this docking experiment ultimately converged on a tight ensemble of conformations. Next, these models were flexibly fit to the nine different conformations observed in the RCT reconstructions. Due to the large conformational changes between the closed and open states, and the limited radius of convergence of our refinement, these models were iteratively fit, fitting first to the most closed state, then to the second most-closed state, and so on.
Refinement of models into each state consisted of a two-step approach: first, models were minimized in torsion space using a differentiable low-resolution “centroid” energy where protein side chains are represented by single interaction centers (Song et al., 2013). Next, all-atom refinement was carried out in Cartesian space using the Rosetta FastRelax protocol (Conway et al., 2014). During the first stage, weak intra-domain Gaussian atom-pair distance constraints were employed to ensure each chain did not move too far from the starting conformation. No additional driving forces were added other than the experimental data. Side chains were truncated at Cβ in the deposited models due to the limited resolution of the reconstructions.
Supplementary Material
Morph of the random conical tilt 3D reconstructions showing LM609 bound to all observed integrin conformations ranging from bent to extended states. The theoretical conformational trajectory shown here was mapped by using LM609 and the αVβ3 integrin headpiece domain as a fiducial during superimposition of all reconstructions.
Highlights.
Crystal Structure of the LM609 Antigen Binding Fragment
Single Particle EM Reveals LM609 Binds to the Headpiece Region in all αVβ3 Integrin Conformations
The LM609 Epitope does not Overlap with the RGD Ligand Binding Pocket
LM609 Sterically Hinders Access of Large Ligands to the αVβ3 RGD Binding Site
Acknowledgments
We thank Ahmed Haouz and members of the crystallogenesis facility at Institut Pasteur for the use of their robotized system, and staff at the SOLEIL synchrotron Proxima I beam line for help with diffraction data collection with the LM609 Fab crystals. We are grateful to John Sumida at the Analytical Biopharmacy Core at the University of Washington for his assistance with surface plasmon resonance studies. Research reported in this publication was supported by the National Institute of General Medical Sciences under Award Number 1R01GM120553 (DV) and T32GM008268 (A.J.B.), a Pew Scholars Award (D.V.), the Raymond and Beverly Sackler Scholars Program in Integrative Biophysics (Z.M.J) and grant DEntry ANR-05-MIIM-012-02 (M.B. and F.A.R.). F.A.R. acknowledges support from CNRS and Institut Pasteur recurrent funding, as well as the Pasteur-Weizmann Servier prize.
Footnotes
Author contributions
A.J.B., Z.M.J, W.N.Z, F.A.R., F.D., M.B. and D.V. designed the research. M.G. contributed key reagents. A.J.B., Z.M.J., F.D., M.B. and D.V. performed the research. A.J.B. and D.V. analyzed the data and wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interest.
Supplemental information includes one table, five figures, and can be found with this article online at http://xxxxxxxxxxxxxxxxxxxxx.xxxxx/xxxx
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Veesler (dveesler@uw.edu).
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical analysis was performed on data obtained in this study.
DATA AND SOFTWARE AVAILABILITY
Accession codes
The crystal structure of the LM609 Fab has been deposited to the protein data bank with accession code 5OPY. The EM maps and atomic models have been deposited to the electron microscopy and protein data banks with accession codes EMD-7011 (bent conformation), EMD-7012 (intermediate conformation), EMD-7013 (extended conformation), PDB 6AVQ (bent conformation), PDB 6AVR (intermediate conformation) and PDB 6AVU (extended conformation).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Backovic M, Johansson DX, Klupp BG, Mettenleiter TC, Persson MA, Rey FA. Efficient method for production of high yields of Fab fragments in Drosophila S2 cells. Protein Eng Des Sel. 2010;23:169–174. doi: 10.1093/protein/gzp088. [DOI] [PubMed] [Google Scholar]
- Backovic M, Krey T. Stable Drosophila Cell Lines: An Alternative Approach to Exogenous Protein Expression. Methods Mol Biol. 2016;1350:349–358. doi: 10.1007/978-1-4939-3043-2_17. [DOI] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charo IF, Nannizzi L, Smith JW, Cheresh DA. The vitronectin receptor alpha v beta 3 binds fibronectin and acts in concert with alpha 5 beta 1 in promoting cellular attachment and spreading on fibronectin. The Journal of cell biology. 1990;111:2795–2800. doi: 10.1083/jcb.111.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xie C, Nishida N, Li Z, Walz T, Springer TA. Requirement of open headpiece conformation for activation of leukocyte integrin alphaXbeta2. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14727–14732. doi: 10.1073/pnas.1008663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheresh DA. Human endothelial cells synthesize and express an Arg-Gly-Asp-directed adhesion receptor involved in attachment to fibrinogen and von Willebrand factor. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:6471–6475. doi: 10.1073/pnas.84.18.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhier F, Le Breton A, Preat V. RGD-based strategies to target alpha(v) beta(3) integrin in cancer therapy and diagnosis. Mol Pharm. 2012;9:2961–2973. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelinger AL, 3rd, Du XP, Plow EF, Ginsberg MH. Monoclonal antibodies to ligand-occupied conformers of integrin alpha IIb beta 3 (glycoprotein IIb-IIIa) alter receptor affinity, specificity, and function. The Journal of biological chemistry. 1991;266:17106–17111. [PubMed] [Google Scholar]
- Garrigues HJ, Rubinchikova YE, Dipersio CM, Rose TM. Integrin alphaVbeta3 Binds to the RGD motif of glycoprotein B of Kaposi’s sarcoma-associated herpesvirus and functions as an RGD-dependent entry receptor. Journal of virology. 2008;82:1570–1580. doi: 10.1128/JVI.01673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. Journal of virology. 1999;73:3951–3959. doi: 10.1128/jvi.73.5.3951-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. Journal of structural biology. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Gramoun A, Shorey S, Bashutski JD, Dixon SJ, Sims SM, Heersche JN, Manolson MF. Effects of Vitaxin, a novel therapeutic in trial for metastatic bone tumors, on osteoclast functions in vitro. J Cell Biochem. 2007;102:341–352. doi: 10.1002/jcb.21296. [DOI] [PubMed] [Google Scholar]
- Gutheil JC, Campbell TN, Pierce PR, Watkins JD, Huse WD, Bodkin DJ, Cheresh DA. Targeted antiangiogenic therapy for cancer using Vitaxin: a humanized monoclonal antibody to the integrin alphavbeta3. Clin Cancer Res. 2000;6:3056–3061. [PubMed] [Google Scholar]
- Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T, Handa M, Takakuwa S, Sato Y, Kawai Y, Ikeda Y, Aiso S. Epitope mapping for monoclonal antibody reveals the activation mechanism for alphaVbeta3 integrin. PloS one. 2013;8:e66096. doi: 10.1371/journal.pone.0066096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- Krey T, d’Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Liu Z, Jia B, Zhao H, Chen X, Wang F. Specific targeting of human integrin alpha(v)beta (3) with (111)In-labeled Abegrin in nude mouse models. Mol Imaging Biol. 2011;13:112–120. doi: 10.1007/s11307-010-0302-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Oliveira dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, Potter CS, Carragher B, Joazeiro CA. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15981–15986. doi: 10.1073/pnas.1413882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam B, Van Agthoven JF, Xiong JP, Alonso JL, Adair BD, Rui X, Anand S, Mehrbod M, Mofrad MR, Burger C, et al. Atomic basis for the species-specific inhibition of alphaV integrins by monoclonal antibody 17E6 is revealed by the crystal structure of alphaVbeta3 ectodomain-17E6 Fab complex. The Journal of biological chemistry. 2014;289:13801–13809. doi: 10.1074/jbc.M113.546929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard M, Odde S, Neamati N. Integrin targeted therapeutics. Theranostics. 2011;1:154–188. doi: 10.7150/thno/v01p0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Mosesson MW. Fibrinogen and fibrin structure and functions. J Thromb Haemost. 2005;3:1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- Posey JA, Khazaeli MB, DelGrosso A, Saleh MN, Lin CY, Huse W, LoBuglio AF. A pilot trial of Vitaxin, a humanized anti-vitronectin receptor (anti alpha v beta 3) antibody in patients with metastatic cancer. Cancer Biother Radiopharm. 2001;16:125–132. doi: 10.1089/108497801300189218. [DOI] [PubMed] [Google Scholar]
- Rader C, Cheresh DA, Barbas CF., 3rd A phage display approach for rapid antibody humanization: designed combinatorial V gene libraries. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8910–8915. doi: 10.1073/pnas.95.15.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka M, Springer TA. Therapeutic antagonists and conformational regulation of integrin function. Nat Rev Drug Discov. 2003;2:703–716. doi: 10.1038/nrd1174. [DOI] [PubMed] [Google Scholar]
- Sidhu SS, Kossiakoff AA. Exploring and designing protein function with restricted diversity. Curr Opin Chem Biol. 2007;11:347–354. doi: 10.1016/j.cbpa.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PL, Nemerow GR. Cell integrins: commonly used receptors for diverse viral pathogens. Trends in microbiology. 2007;15:500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–511. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Triantafilou K, Triantafilou M, Takada Y, Fernandez N. Human parechovirus 1 utilizes integrins alphavbeta3 and alphavbeta1 as receptors. Journal of virology. 2000;74:5856–5862. doi: 10.1128/jvi.74.13.5856-5862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Agthoven JF, Xiong JP, Alonso JL, Rui X, Adair BD, Goodman SL, Arnaout MA. Structural basis for pure antagonism of integrin alphaVbeta3 by a high-affinity form of fibronectin. Nature structural & molecular biology. 2014;21:383–388. doi: 10.1038/nsmb.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeravagu A, Liu Z, Niu G, Chen K, Jia B, Cai W, Jin C, Hsu AR, Connolly AJ, Tse V, et al. Integrin alphavbeta3-targeted radioimmunotherapy of glioblastoma multiforme. Clin Cancer Res. 2008;14:7330–7339. doi: 10.1158/1078-0432.CCR-08-0797. [DOI] [PubMed] [Google Scholar]
- Veesler D, Cupelli K, Burger M, Graber P, Stehle T, Johnson JE. Single-particle EM reveals plasticity of interactions between the adenovirus penton base and integrin alphaVbeta3. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8815–8819. doi: 10.1073/pnas.1404575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. Journal of structural biology. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Beuerlein G, Nie Y, Smith H, Lee BA, Hensler M, Huse WD, Watkins JD. Stepwise in vitro affinity maturation of Vitaxin, an alphav beta3-specific humanized mAb. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6037–6042. doi: 10.1073/pnas.95.11.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Avila-Sakar A, Kim J, Booth DS, Greenberg CH, Rossi A, Liao M, Li X, Alian A, Griner SL, et al. Fabs enable single particle cryoEM studies of small proteins. Structure. 2012;20:582–592. doi: 10.1016/j.str.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Mahalingham B, Alonso JL, Borrelli LA, Rui X, Anand S, Hyman BT, Rysiok T, Muller-Pompalla D, Goodman SL, et al. Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment. The Journal of cell biology. 2009;186:589–600. doi: 10.1083/jcb.200905085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. The Journal of cell biology. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Hamer DH, Hendrickson WA, Sattentau QJ, Kwong PD. Interfacial metal and antibody recognition. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14575–14580. doi: 10.1073/pnas.0507267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Molecular cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morph of the random conical tilt 3D reconstructions showing LM609 bound to all observed integrin conformations ranging from bent to extended states. The theoretical conformational trajectory shown here was mapped by using LM609 and the αVβ3 integrin headpiece domain as a fiducial during superimposition of all reconstructions.