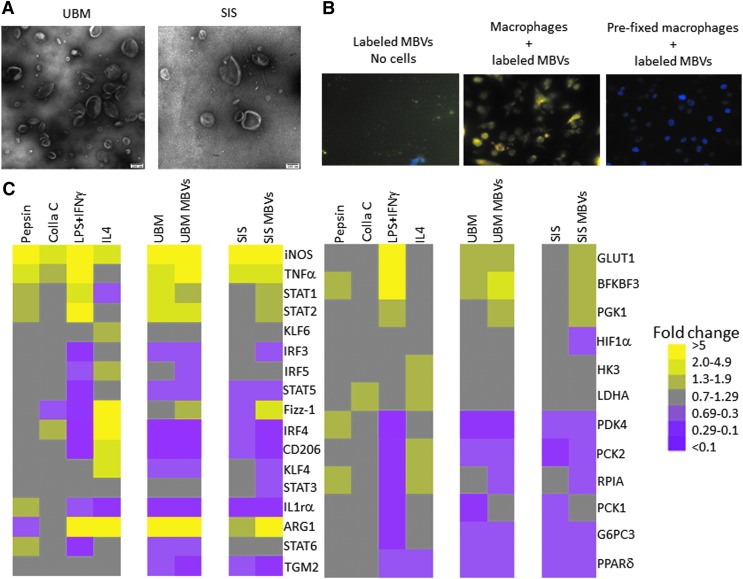
FIG. 1.
MBV imaging and gene expression signature of MBV-treated macrophages. (A) MBV isolated using proteinase-K digestion were visualized by transmission electron microscopy at 100,000 × magnification. (B) Following exposure of BMDMs (counterstained with DAPI) to MBVs whose nucleic acid content was labeled with acridine orange, the cells were visualized using fluorescence microscopy at 200 × magnification. MBV are seen within the cytosol of cells after a 2-h incubation. (C) Gene expression analysis of cells exposed to either ECM or their respective MBVs was evaluated using qPCR. Results are presented in a heatmap form that was generated using Treeview software; all fold changes are with respect to media control (N = 3). Scale bar scoring system is demonstrated as follows: less than 0.1-fold change (darkest purple), 0.1–0.29-fold change (intermediate purple), 0.3–0.69-fold change (light purple), 0.7–1.29-fold change (gray), 1.3–1.9-fold change (light yellow), 2.0–4.9-fold change (intermediate yellow), greater than 5.0-fold change (bright yellow). Supplementary Table S1 indicates significant differences between media control and treated macrophages as determined using t-tests with p < 0.05 considered significant. BMDM, bone marrow-derived macrophages; ECM, extracellular matrix; DAPI, 4’6-diamidino-2-phenylindole; MBV, matrix-bound nanovesicles.