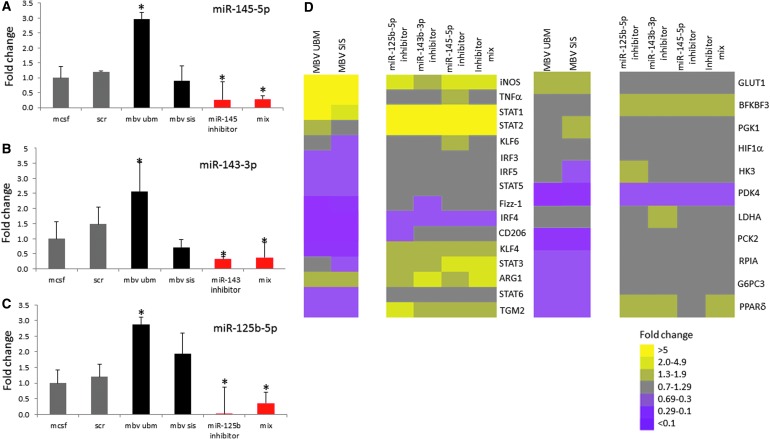
FIG. 4.
miRNA inhibition reverses gene expression patterns compared to MBV-exposed BMDM. Macrophage miRNA inhibition. (A–C) Selective inhibition of specific miRNAs, miR-145-5p, miR-145-3p, and miR-125-b-5p using 50 nM of inhibitor for each. Relative abundance of miRNA levels following inhibition was determined by TaqMan miRNA qPCR assays. Values: mean ± standard deviation, N = 3, *p < 0.05 by Student's t-test. Data represents fold change in comparison to M0. (D) Gene expression analysis of cells exposed to MBVs, or transfected with scrambled control miRNA inhibitor, mmu-miR-154-5p inhibitor, mmu-miR-143-3p inhibitor, mmu-miR-125b-5p inhibitor, or a combination of all three inhibitors was evaluated using qPCR. Results are presented in a heatmap form that was generated using Treeview software; all fold changes are with respect to media control. Scale bar scoring system is demonstrated as follows: less than 0.1-fold change (darkest purple), 0.1–0.29-fold change (intermediate purple), 0.3–0.69-fold change (light purple), 0.7–1.29-fold change (gray), 1.3–1.9-fold change (light yellow), 2.0–4.9-fold change (intermediate yellow), and greater than 5.0-fold change (bright yellow). Supplementary Table S3 indicates significant differences between media control and treated macrophages as determined using t-tests with p < 0.05 considered significant.