Fig. 3.
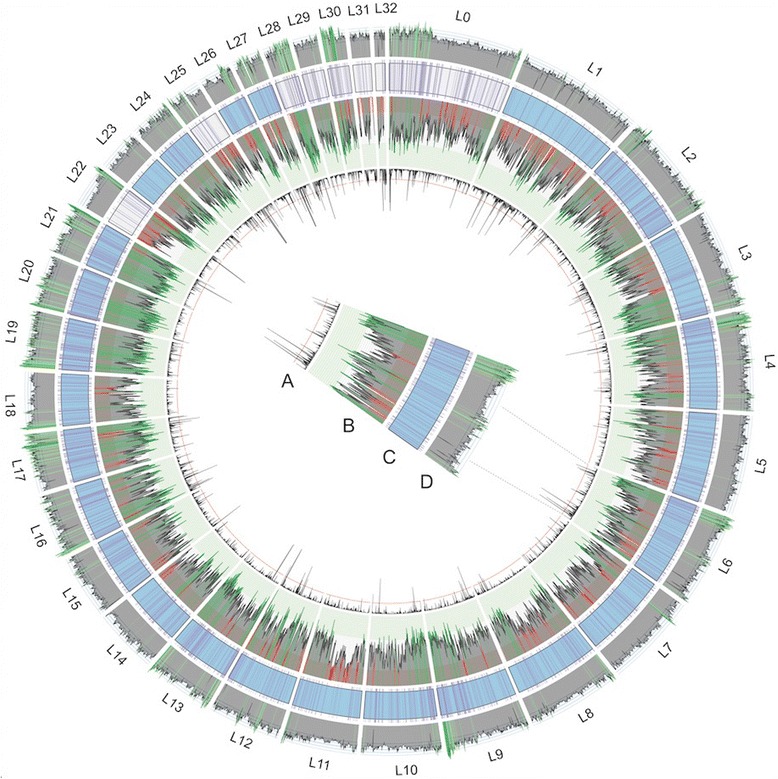
Mouse lemur 3.0 assembly. Circos plots were calculated with a sliding window of 500 kb. a Linear plot of percent of gaps encoded as N’s, plotted inward, where the red horizontal line is 25%. b Histogram of BNG physical map coverage across the scaffold, plotted with three horizontally shaded zones that match the data’s quartiles: 35× coverage and below is red (less than Q1), 35–56× coverage is grey (Q1–Q3), and 56× coverage and above is green (greater than Q3). c Lachesis scaffolds arranged according to length (in base pairs). Blue colored scaffolds represent those assigned to mouse lemur chromosomes (see Fig. 5) and white scaffolds are undetermined. Purple hashes identify regions containing the complete single copy genes (n = 2628) according to BUSCO analysis. d Histogram of percent of bases that are G + C across the genome. Genome-wide average is 40.98%, regions shaded light green are at least 47.5%, and regions shaded dark green are at least 55% G + C content
