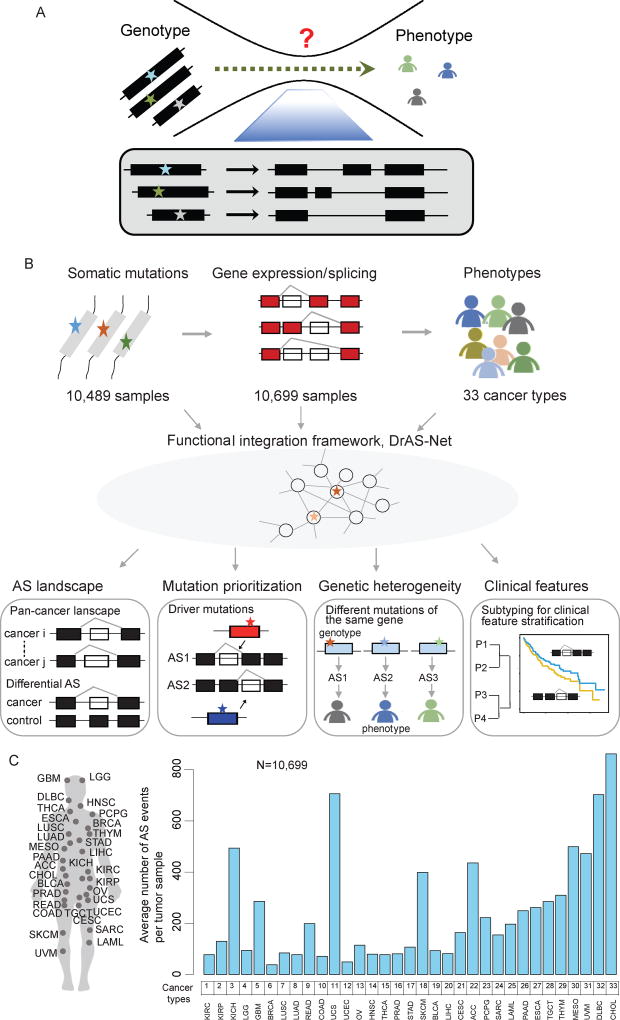
Figure 1. Systematic Characterization of Mutation-Mediated Alternative Splicing Events across 33 Cancer Types.
(A) Alternative splicing (AS) underlies the complexity of genotype-phenotype relationships.
(B) Flowchart of the mutation-mediated alternative splicing (AS) analysis in cancer. Genome-wide mutational profiles of 10,489 samples and AS data from 10,699 samples across 33 types of cancer are integrated into functional networks. Four types of analyses are shown: I) Identification of genome-wide AS alternations in each type of cancer. Differential AS events are identified as cancer-specific splicing compared to controls; II) Prioritization of driver somatic mutations based on the functional networks. The functional importance of mutations is evaluated; III) Proposed mutation-AS model to explain principles of genetic heterogeneity; IV) Clustering analysis based on AS to identify cancer subtypes with distinct clinical features. P, patient.
(C) The average number of AS events per tumor detected in each cancer type from a total of 10,699 samples.
See also Table S1.