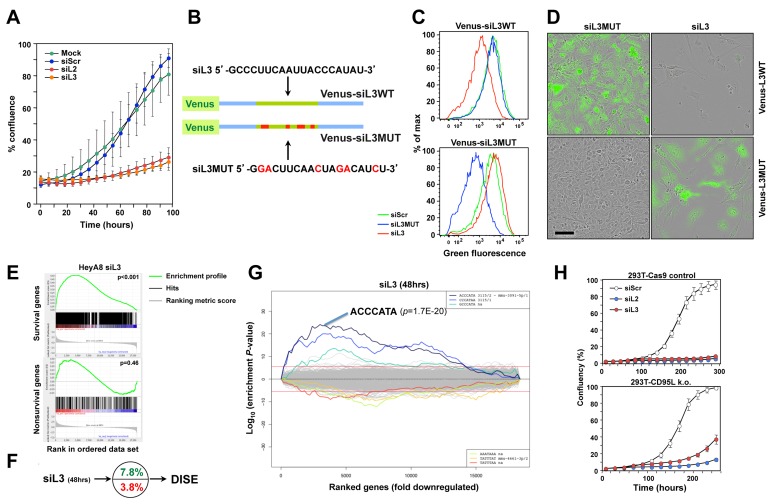
Figure 3. CD95L derived siL2 and siL3 knockdown CD95L and induce DISE by targeting critical survival genes.
A. Specific targeting of a minisensor containing Venus fused to a CD95L minigene comprised of the 50 nt surrounding the siL3 target site in CD95L. Cells were either mock transfected (Lipofectamine only) or transfected with either 5 nM siScr, siL2 or siL3. Shown is change in percent confluency over time when cells were plated at 3000 cells per well in a 48-well plate 24 hours after transfection. B. Schematic showing the design of the wt Venus-siL3 sensor and a sensor carrying a mutated siL3 targeted site. Sequences of targeting siRNAs are given. C. FACS analysis of HeyA8 cells stably expressing either the wt or the mutant Venus-siL3 sensor (see B) 24 after transfection with 10 nM of either siScr, siL3WT or siL3MUT. D. Composite images of phase contrast and green fluorescence of HeyA8 cells expressing either Venus-siL3WT or Venus-siL3MUT sensor 6 days after transfection with 10 nM of either siL3 or siL3MUT. Scale bar = 150 µm. E. Gene set enrichment analysis for a group of 1846 survival genes (top panel) and 416 nonsurvival genes (bottom panel) (see [9]) after introducing siL3 in HeyA8 cells. siScr served as control. p-values indicate the significance of enrichment. F. Preferential downregulation of survival genes in cells transfected with siL3 as in E. G. SYLAMER analysis of HeyA8 cells 50 hrs after transfection with 25 nM siL3. H. Change in confluency over time of a mix of three 293T-Cas9 clones compared to a mix of two complete 293T CD95L deletion clones after transfection with 25 nM siScr, siL2 or siL3 (all from Dharmacon, IDT oligonucleotides gave very similar results (data not shown)).