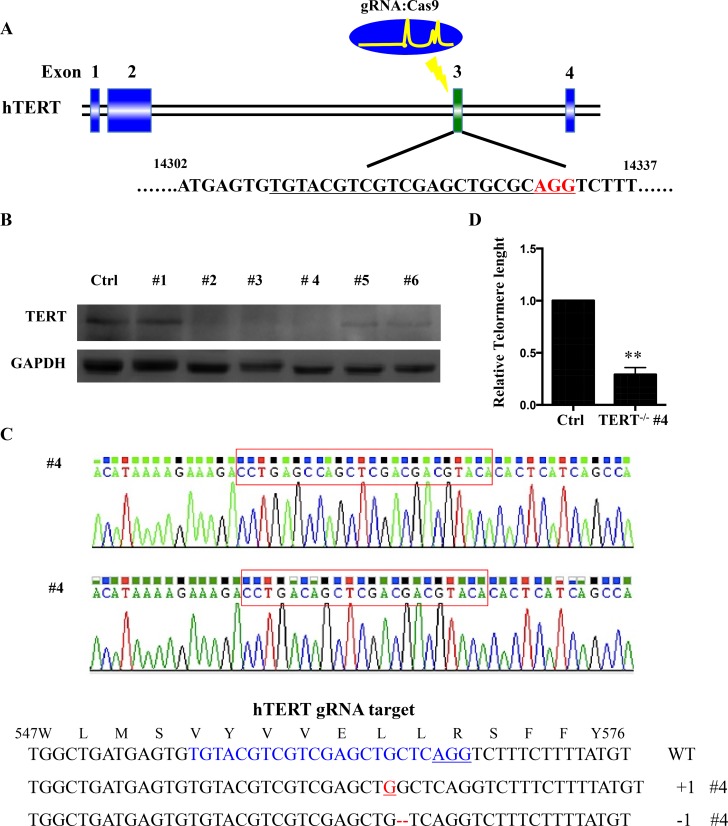
Figure 2. hTERT knockout cells generated using CRISPR-Cas system.
(A) Schematic representation of the genomic DNA structure of hTERT with exons numbered. The sequence including AGG PAM targeted by gRNA in CRISPR-Cas9 system is shown in red. (B) Western blot analysis of various knockout cell clones to evaluate the expression of hTERT in squamous cell carcinoma. GAPDH was used as loading control. (C) Clone #4 had the highest degree of hTERT reduction and was sequenced and aligned with the wild type sequence (+: inserted bases; −: deleted bases). (D) Relative telomere lengths were detected by RT–PCR between hTERT knockout cell line and the control.