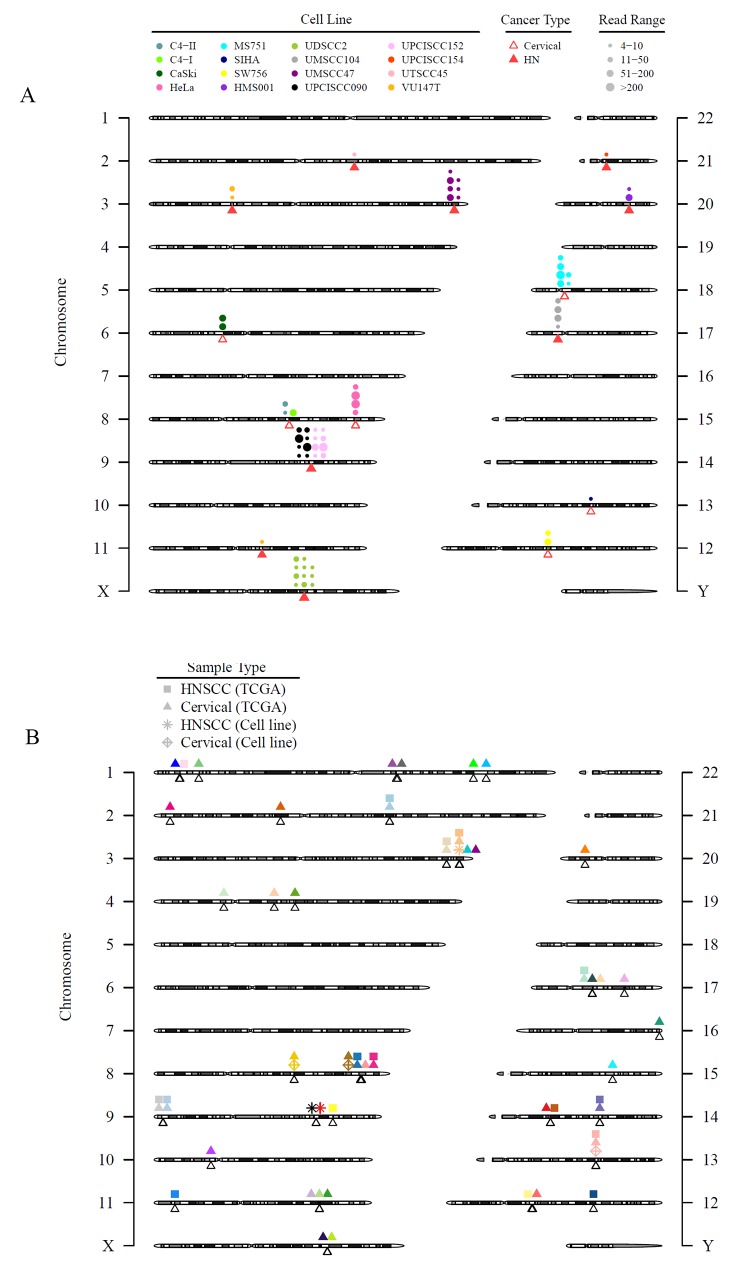
Figure 3. HPV integration sites in HNSCC and CESC cell lines and tissues are diverse with few areas of recurrent integration.
Viral integration sites for all HPV-positive cell lines alone (A) and combined with the TCGA HNSCC and CESC tumors (B). (A) Triangles point to the exact integration locations and may overlap. Each circle represents an integration event; circles that are close to each other may be shifted for better visualization. The size of the circles represents discordant read pairs in different ranges. A read pair was reported as discordant if the paired-end reads were uniquely mapped with one end to a human chromosome and the other to the virus chromosome. Discordant read pairs are evidence of HPV integration. Larger circles indicate that more discordant read pairs were mapped to that integration site and, therefore, provide stronger evidence for integration. (B) Unfilled triangles point to the exact integration event and may overlap. The shapes of the symbols refer to different sample types or sources. Each color represents a gene, but the location of the symbols may be shifted slightly for better visualization. Only genes with HPV integration in 2 or more patient tumor samples in TCGA or cell lines were plotted.