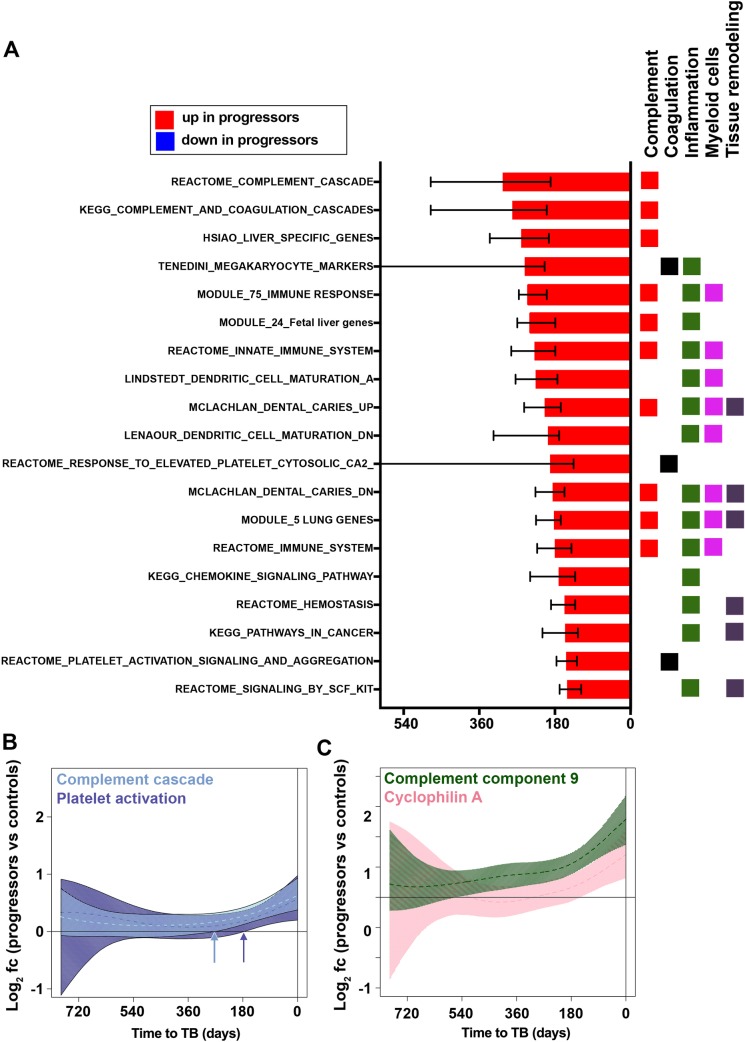
Fig 3. Sequential changes in plasma proteins during progression from M.tb infection to TB disease.
(A) Gene modules, pre-defined by Reactome, KEGG and MSIGDB, and matched to the corresponding protein found to be significantly enriched in plasma from progressors, compared with controls, and ranked in descending order according to median deviation time points (indicated by bars) of proteins differentially abundant between progressors and controls. Data from 36 progressors and 104 controls were included in the analysis. Error bars denote IQR of median deviation time points of differentially abundant plasma proteins within each gene module. Assignment of each module to known immunological responses or processes or cellular subsets, according to differentially abundant proteins, is indicated by the colored squares. The full list of significantly enriched modules is in S5 Table. (B) Kinetics of complement cascade and platelet activation protein modules. Module kinetics during progression were modeled as non-linear splines (dashed lines) and 99% CI (shaded areas) were computed by performing 2000 spline fitting iterations after bootstrap resampling from the full dataset. Arrows indicate the time before TB diagnosis at which the 99% CI deviates from zero for the two modules. (C) Kinetics of individual proteins representing the complement cascade (complement component 9) and platelet activation (cyclophilin A) protein modules, modeled as non-linear splines and 99% CI.