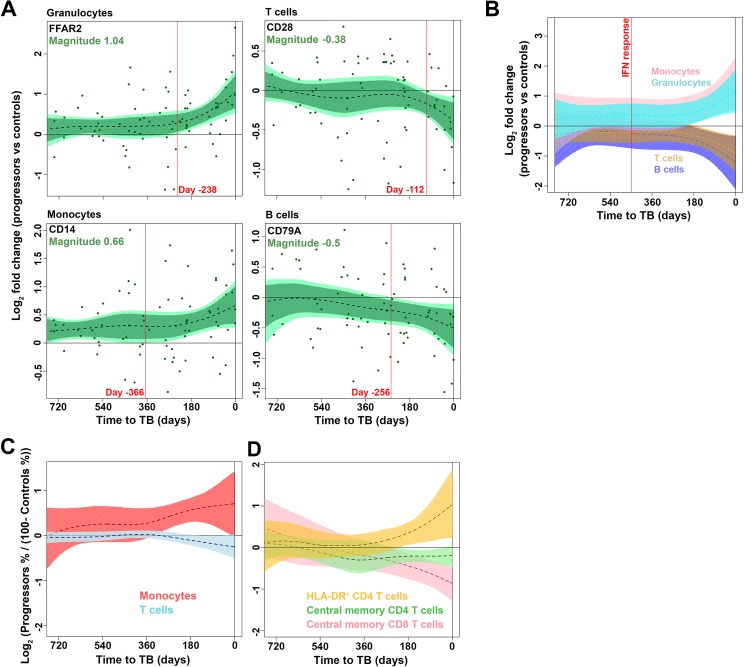
Fig 4. Changes in proportions of peripheral blood cell subsets during progression from infection to TB disease.
(A) Kinetics of mRNA expression, expressed as log2 fold change between bin-matched progressors and controls and modeled as non-linear splines (dotted lines) for genes representing granulocytes, monocytes, T cells and B cells. Light green shading represents 99% CI and dark green shading 95% CI for the temporal trends, computed by performing 2000 spline fitting iterations after bootstrap resampling from the full dataset. The magnitude for each gene, representing the log2 fold change at TB diagnosis, is shown in green text. The deviation time, calculated as the time point at which the 99% CI deviates from a log2 fold change of 0, is indicated in red text. Data from 38 progressors and 104 controls were included in the analysis. (B) Temporal trends of gene modules representing granulocytes (genes with significant kinetic response from BIOCARTA_GRANULOCYTES_PATHWAY), monocytes (genes from M11.0_enriched in monocytes (II)), T cells (genes from M4.1_T-cells, M6.15_T-cells, M7.1_T cell activation (I) and M7.4_T cell activation (III)) and B cells (genes from M4.10_B-cells, M47.0_enriched in B cells (I), M47.1_enriched in B cells (II) and M69_enriched in B cells (VI)), modeled as non-linear splines during progression. The deviation time for the interferon module shown in Fig 3B is denoted by the vertical red line. (C) Temporal trends of relative mean (dotted lines) proportions of monocytes and T cells (D) or activated HLA-DR+ CD4 T cells, CCR7+CD45RA- central memory CD4 or CD8 T cells, measured by flow cytometry from cryopreserved PBMC and modeled as non-linear splines during progression. Shown are log2 cell proportions for progressors relative to bin-matched controls. Data from 33 progressors and 71 controls were included in the analysis. Shading denotes 99% CI computed by performing 2000 spline fitting iterations after bootstrap resampling from the full dataset.