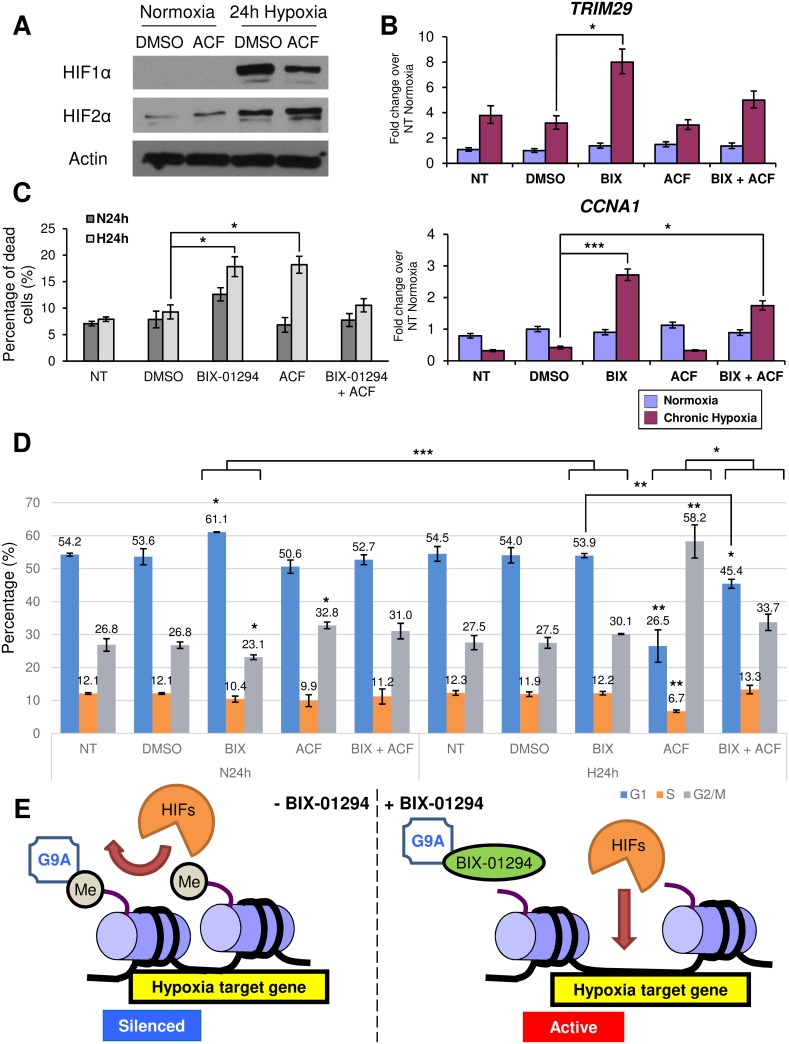
Fig 7. HIF inhibition by ACF antagonizes the response of de novo hypoxia targets derepressed by G9A inhibition to chronic hypoxia.
(A) Western blots show protein levels of HIF1α and HIF2α in MCF-7 cells after 24 hours under the indicated treatments. Actin was used as the loading control. (B) Fold change in gene expression of TRIM29 and CCNA1 in MCF-7 cells treated with 6 μM BIX-01294 (BIX) and/or 0.6 μM ACF compared to the NT and DMSO controls in normoxia (blue) and 24 hours chronic hypoxia (magenta). Gene expression levels were normalized against the housekeeping reference gene EEF1G and fold change was calculated against the average of the NT controls in normoxia. Error bars indicate SEM for n = 9 replicates. (C) Bar chart shows percentage of dead cells staining positive for trypan blue following treatment of MCF-7 cells with the compounds as indicated under 24 hours normoxia (N24h) and 24 hours chronic hypoxia (H24h). Error bars denote SEM for n = 3 replicates. (D) Cell cycle analysis showing the distribution of MCF-7 cells in different cell cycle phases. Bar chart indicates the percentage of cells in the G1 (blue), S (orange) and G2/M (grey) phases of cell cycle out of the total number of cells for each treatment (NT, untreated; DMSO, 0.1% DMSO; BIX, 0.1% DMSO + 6 μM BIX-01294; ACF, 0.1% DMSO + 0.6 μM ACF; N24h, 24 hours normoxia; H24h, 24 hours chronic hypoxia). Error bars indicate SEM for n = 3 replicates. (E) Schema showing the effect of G9A inhibition by BIX-01294 on hypoxia target gene activation. In the absence of BIX-01294 (left), G9A methylates (Me) H3K9, causing the chromatin to maintain a closed conformation characteristic of silenced genes and preventing the HIFs from transcriptionally regulating their target genes. With BIX-01294 (right), G9A is inhibited and unable to methylate H3K9, allowing the chromatin structure to open and the HIFs to regulate these derepressed hypoxia target genes.