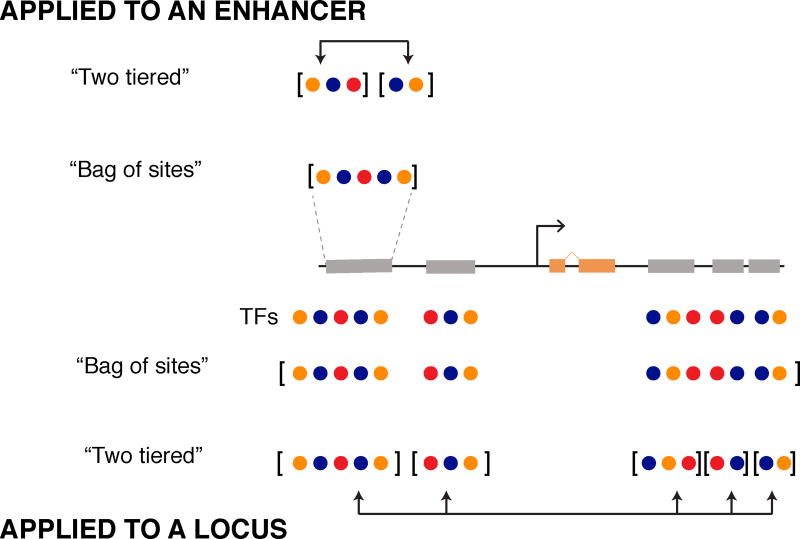
Figure 6. A “two-tiered” mechanism may define and integrate sub-modules of regulatory sequence at the level of single enhancers and entire loci.
(Top) Enhancer sequences contain binding sites for different TFs that function by activating or repressing their target gene. Enhancer-level models capture each TF input independently, representing the enhancer as one “bag of sites”. Two-tier models, such as GEMSTAT-GL, can also be applied to enhancer-length sequences by first separating TF inputs into multiple regulatory segments and then integrating their weighted output to predict expression. (Bottom) Two-tier and enhancer-level models can be applied to an entire locus. Enhancer-level models consider TF binding across the locus as a large “bag of sites”, without considering individual enhancers as separate regulatory entities. We can also apply the two-tiered model to a gene locus. This approach first subdivides the regulatory sequence around a gene into smaller modules and then integrates the regulatory information from each module to predict expression.