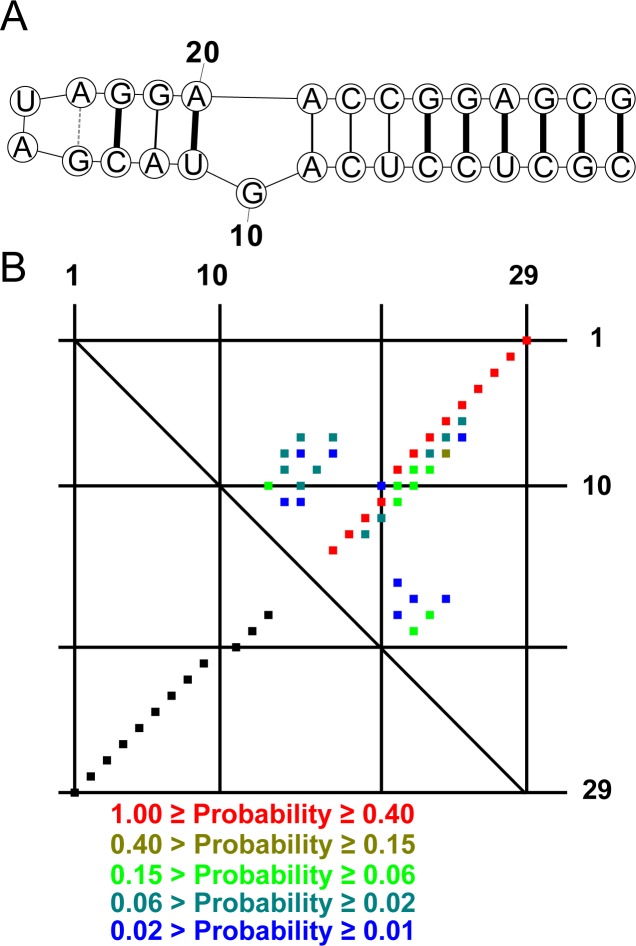
Fig 1. Prediction of extended secondary structure with CycleFold.
(A) A predicted structure for a Sarcin-Ricin loop sequence form Rattus norvegicus [50] using CycleFold with the MFE algorithm. Correctly predicted canonical pairs are drawn with heavy black lines, correctly predicted non-canonical pairs are light black lines, and the incorrectly predicted non-canonical pair is shown with a gray dashed line. The G-A pair at the base of the tetraloop is not present in the reference structure because the 3’ A is not stacked on the subsequent G, but is instead in contact with a protein, Restrictocin. (B) The probability dot plot calculated using Cyclefold with the partition function algorithm. The upper right triangle shows pairs with estimated probabilities > 0.01, color-coded by pairing probability. The lower left triangle shows the pairs that are present in the reference structure. Each dot represents a single base pair, and nucleotide index (starting with 1 at the 5’ end) is shown along the x and y axes.