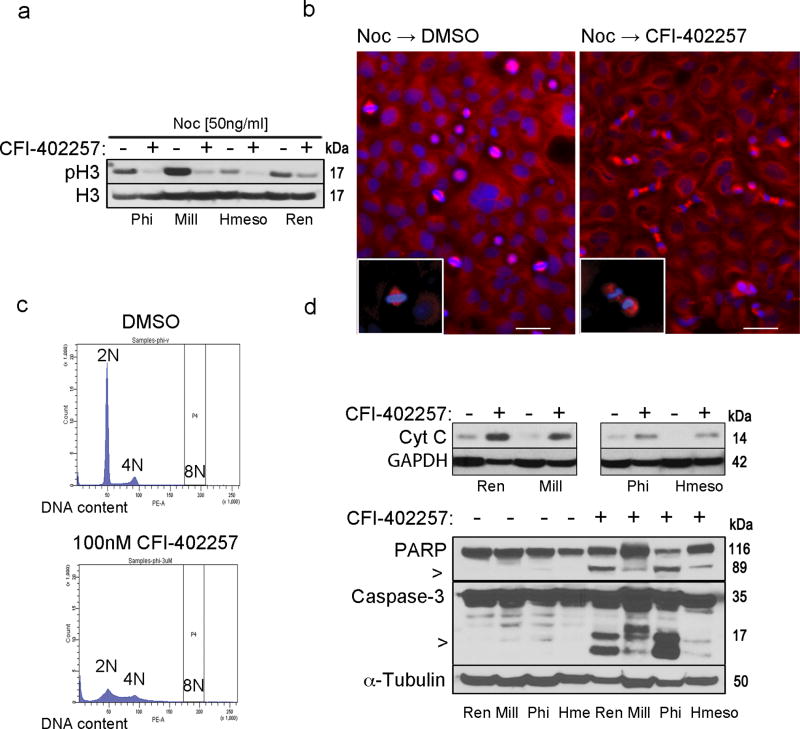
Fig.2. CFI-402257 induces mitotic checkpoint bypass, defects in mitotic division and mitochondrial death of MM cells.
Ren, Mill, Phi and Hmeso cells were treated with 50 ng/ml nocodazole (Noc) (Sigma-Aldrich, MO, US) for 10 hr and then released from the G2/M arrest in full media containing 100 nM CFI-402257 or DMSO. (a) After 3 hr cells were collected and the immunoblot analysis of total cell lysates was performed with phospho-H3 and total H3 antibody as the loading control. Picture is a representative of two independent experiments performed with pSer10H3 and pSer28H3 (#9701 and #9713, Cell Signaling, US) antibodies. (b) After 30 min cells were stained for mitotic spindle (α-tubulin: T6199, Sigma-Aldrich, MO, US) and nuclei (DAPI). Picture magnification is 10×. Bar: 200 µm. Insets show 40× magnification of the cell in metaphase and telophase (boxed regions). (c) Phi cells were treated with 100 nM CFI-402257 or DMSO for 72 hr, and then ethanol-fixed and stained with propidium iodide for the cytofluorometric analysis of DNA content. Data are representative cell cycle histograms. Experiment was repeated in other MM cell lines with similar results. (d) Phi and Hmeso cells were treated with 100 nM CFI-402257 or DMSO. To assess Cyt c release (top panel) after 72 hr of treatment, fractionation was performed by incubation of harvested cells in lysis buffer (20 mM HEPES-KOH pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM bisodium EDTA, 1 mM EGTA, sucrose) and aspiration through a 22-gauge needle followed by centrifugation at 1000g for 5min at 4C to separate nuclei. The supernatant was centrifuged at 10 000g for 20 min in 4C to separate mitochondria. The cytoplasmic fraction was immunoblotted and Cyt c (#556432, BD, CA, US) was detected. After 5 days of treatment, total lysates of Ren, Mill, Phi and Hmeso were collected and immunoblotted. Caspase-3 and PARP (#9662 and #9542, Cell Signaling, US) were detected (bottom panel), α-tubulin and GAPDH served as a loading control. All the data are representative of two independent experiments. For space constraints, ‘Hme’ stands for ‘Hmeso’ MM cell line.