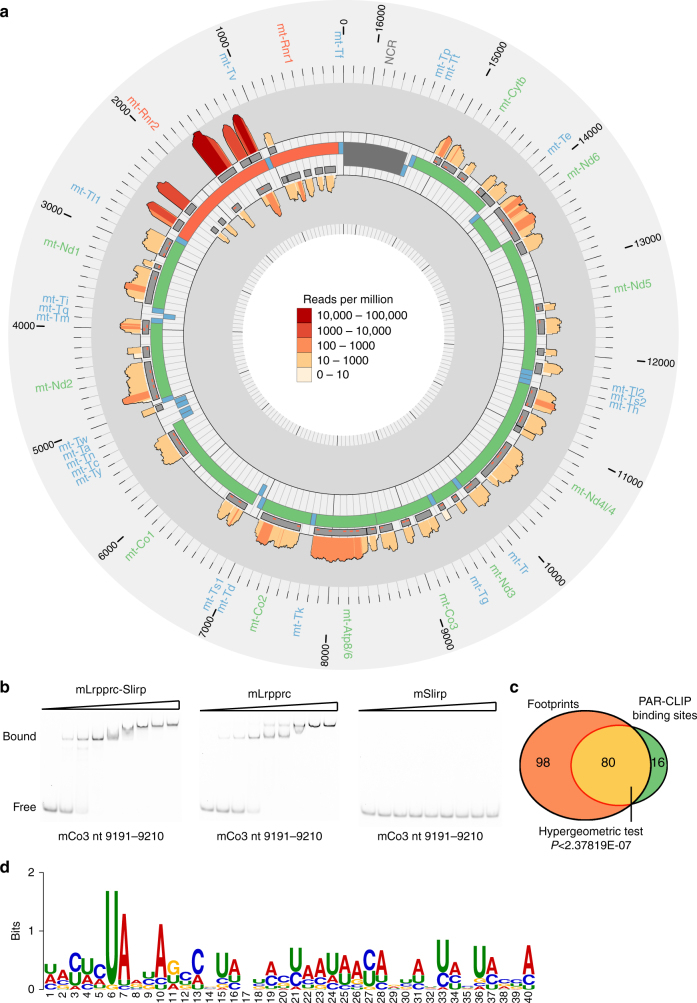
Fig. 2.
PAR-CLIP analysis of LRPPRC-binding sites. a Circular representation of the mitochondrial genome (centre track) displaying the PAR-CLIP-binding site regions (interior tracks) and normalized coverage (log10 (reads per million); scale: 1–100,000) across positions with at least a 95% posterior probability of being cross-linked (exterior tracks), as identified by BMix50. b RNA EMSA of the LRPPRC–SLIRP complex, LRPPRC only or SLIRP only incubated with a RNA probe identified as a strong LRPPRC–SLIRP-binding motif in vivo. c Schematic of shared binding sites among footprinting and PAR-CLIP data sets; the significance (P) of the overlap is indicated. d A sequence logo shows a predicted consensus binding motif for LRPPRC based on the binding sites that overlap between footprinting and PAR-CLIP data sets