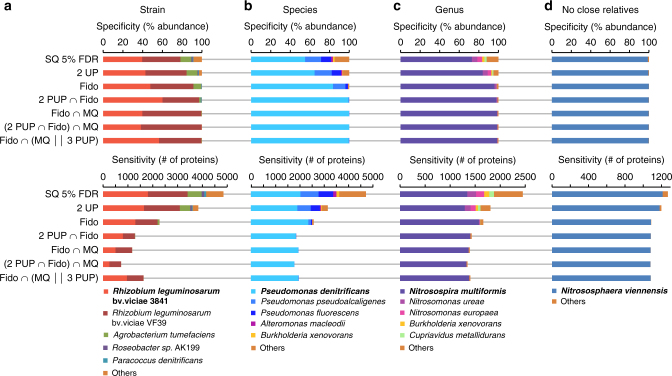
Fig. 2.
Specificity and sensitivity of protein identification with different protein inference strategies. The proteomics data for four pure cultures (a R. leg. bv. viciae 3841, b P. denitrificans, c N. multiformis, d N. viennensis) were analyzed in separate runs. For all cultures, proteins were identified with the same simulated metagenomic database containing 30 different species, which included other organisms related at the strain (a), species (b), or genus level (c). For d, no closely related organisms were included. Upper panels show specificity (% abundance of proteins attributed to each organism) and lower panels sensitivity (# of proteins identified). More stringent identification strategies improve specificity at the expense of sensitivity. Protein inference strategies: SQ 5% FDR; SEQUEST search filtered for 5% false discovery rate (FDR) using standard Target-Decoy strategy (implemented as Protein FDR Validator Node in Proteome Discoverer). 2 UP; same as previous, but only the subset of proteins identified with at least two unique peptides (UP). Fido; SEQUEST-Fido results filtered at 5% FDR based on protein q-value. 2 PUP ∩ Fido; same as previous, but only the subset of proteins identified with at least two protein unique peptides (PUP). Fido ∩ MQ; Only proteins identified both by Sequest-Fido (FDR 5%) and MaxQuant (1% protein FDR, at least 2 razor + unique peptides). (2 PUP ∩ SQ Fido) ∩ MQ; Same as previous, but for SEQUEST-Fido only the subset of proteins identified with at least two protein unique peptides. Fido ∩ (MQ || 3 PUP); Only proteins that were identified both by SEQUEST-Fido and MaxQuant. Additionally, SEQUEST-Fido identified proteins were retained even if they were not identified by MaxQuant if they had at least three protein unique peptides