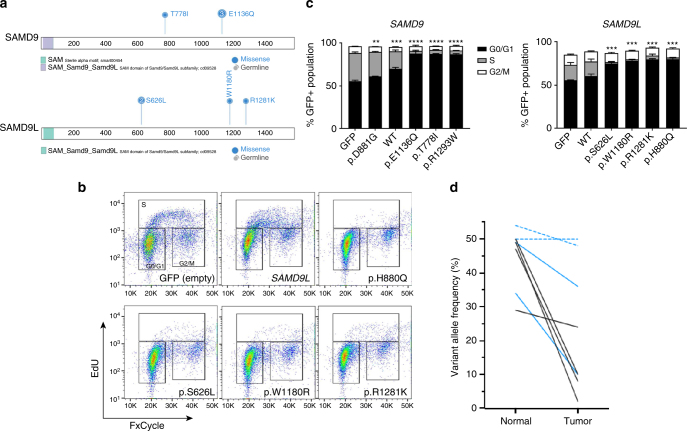
Fig. 4.
Gain-of-function mutations in SAMD9 and SAMD9L decrease cell proliferation and inhibit cycle progression. a Schematic showing the protein structure of SAMD9 and SAMD9L. b Flow cytometry plots (FxCycle-total DNA content vs. EdU incorporation) showing the cell cycle inhibiting effects of gain-of-function SAMD9L mutations. SAMD9L p.H880Q (positive control) is a gain-of-function mutation previously reported in Ataxia-Pancytopenia syndrome21. c EdU incorporation assay showing that gain-of-function SAMD9/SAMD9L mutations inhibit cells from progressing through the cell cycle as depicted by the relative absence of cells in S-phase. SAMD9 p.R1293W (positive control) is a previously reported gain-of-function mutation in MIRAGE syndrome19. SAMD9 p.D881G is a common SNP that is not predicted to be pathogenic. **: p < 0.01, ***: p < 0.001, ****: p < 0.0001 (student’s t-test). Error bars indicate standard deviation. Data in B & C are representative of 2–3 experiments completed in triplicate. d VAF plot, obtained from targeted deep sequencing, showing the preferential loss (decrease in VAF) of the SAMD9/SAMD9L mutations in the tumor population. Black lines: SAMD9; Blue lines: SAMD9L; dashed lines indicate cases where a subclonal (3/20 metaphases) del(7) was detected only by conventional karyotyping