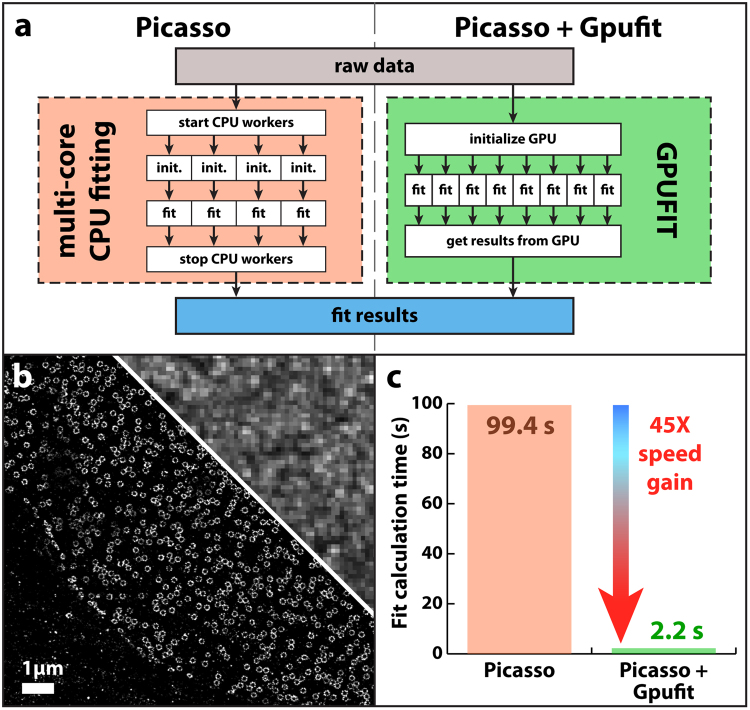
Figure 4.
Accelerated STORM analysis in Picasso. (a) Schematic diagram comparing the multi-core CPU-based fitting of Picasso (left) vs. parallelized GPU-based fitting with Gpufit (right). Picasso uses multiple cores (“workers”) of the CPU to simultaneously compute the fits. We replaced this section of the Picasso source code with a call to Gpufit, which has a greater capacity for parallelization via the GPU. (b) Super-resolution (STORM) image of nuclear pore complexes in eukaryotic cells. The protein GP210 was labeled with a fluorescent marker (Alexa Fluor 647), and the sample was imaged on a STORM microscope (see Supplementary Information). The conventional epi-fluorescence image is shown in the top right corner, to provide a resolution comparison. Data processing was performed in Picasso, requiring 3.6 million individual curve fitting operations. (c) Execution time of the curve fitting process, comparing the original (published) Picasso software vs. the modified Picasso software which includes Gpufit. The processing time was reduced significantly, by a factor of 45, when Gpufit was used to handle curve fitting tasks within the Picasso application. For these measurements, the data was fit to a symmetric 2D Gaussian function (Picasso default) using the unweighted least squares estimator, and the fit size was 7 × 7 pixels. All calculations were run on an NVIDIA GeForce GTX 1080 GPU and an Intel Core i7 5820 K CPU (3.3 GHz). For detailed test conditions, see the Supplementary Information.