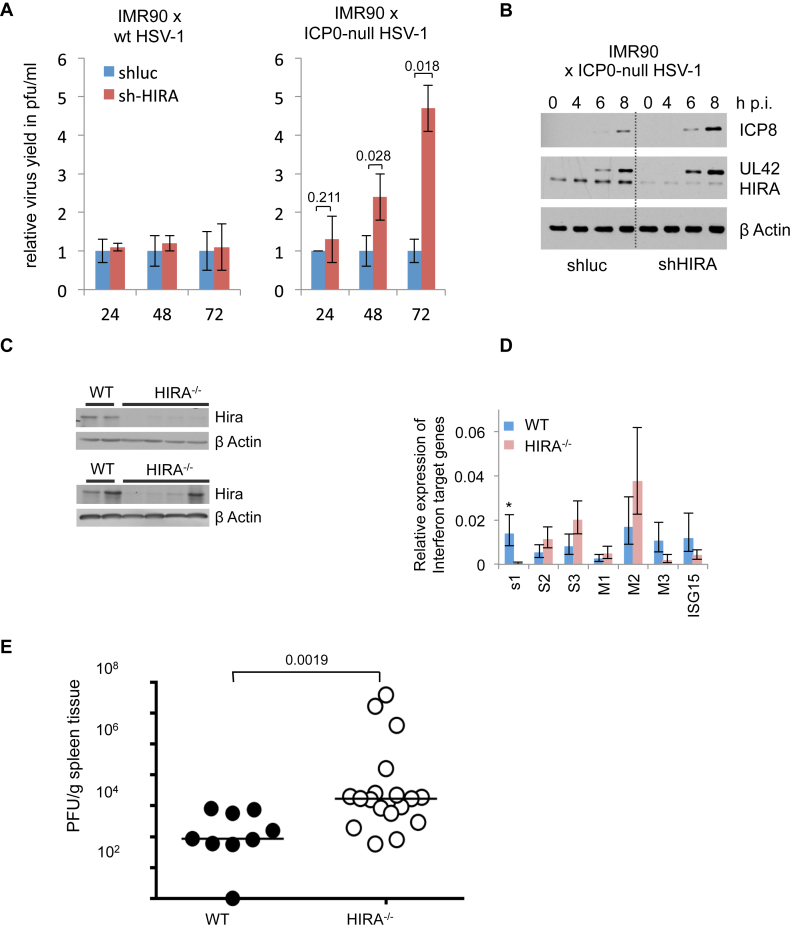
Figure 4.
HIRA contributes to efficient suppression of viral infection. (A) Virus yield from HIRA-depleted and control IMR90 cells infected at MOI 0.01 with ICP0-null HSV-1 mutant dl1403 CMV lacZ or wt HSV-1 variant in1863. Supernatant was harvested at indicated times post infection (h p.i.) and virus titres determined by plaque assay. Data are mean +/- SD (error bars) (n = 3 biological repeats) with indicated P values. (B) HIRA-depleted and control IMR90 cells infected with ICP0-null HSV-1 mutant dl1403 at MOI 2.0. Lysates were harvested and processed at indicated time points post infection (h p.i.) (C–E) Control (CAGG-Cre-ER, WT, +tamoxifen) or Hira-deficient (CAGG-Cre-ER, Hirafl/fl, +tamoxifen) mice were infected with MCMV and spleen harvested 4 days later. Each spleen was divided into three pieces for downstream analysis. (C) Western blot analysis of WT or HIRA−/− animals showing knock out of Hira. Shown are representative western blot results from two different gels with 4 WT and 8 HIRA−/-mice. (D) mRNA abundance of IFN-β target genes by qRT-PCR. Bar chart displays mean of each IFN-β target gene mRNA abundance in WT mice compared to HIRA−/-mice, normalized to β-actin as housekeeping control. Data are mean ± SEM (error bars) (n = 4 for WT mice and n = 8 for HIRA−/-mice). *P < 0.05 (OAS1). P > 0.05 for the other six genes (OAS2, OAS3, IFITM1, IFITM2, IFITM3 and ISG15). (E) Plaque forming units measured per gram of tissue of WT or HIRA−/− mice. P-value assessed by Mann–Whitney-U test (n = 9 for WT mice and n = 18 for HIRA−/-mice).