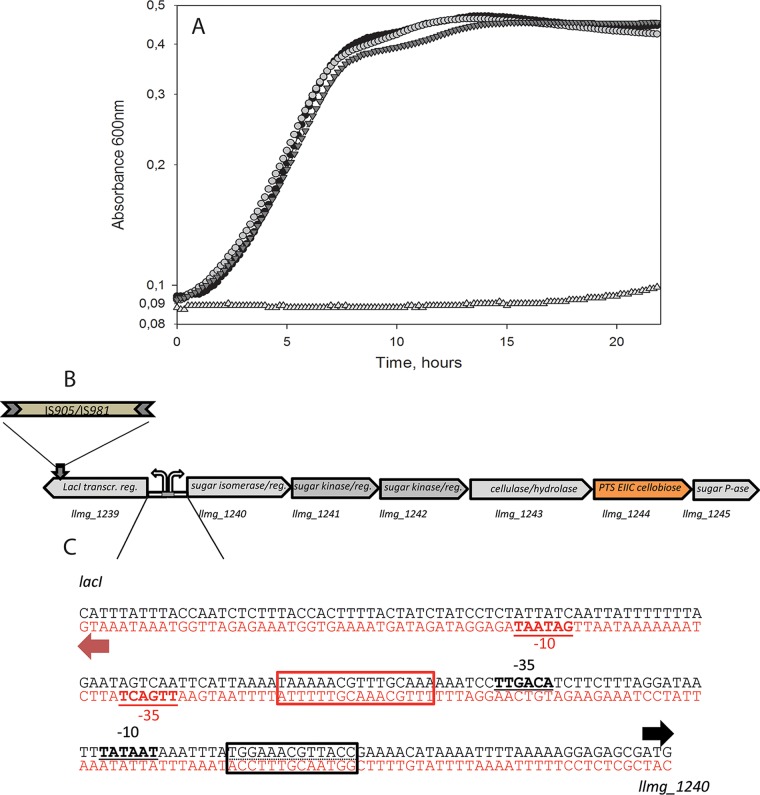
FIG 2.
Integration of IS/GFP into llmg_1239 and growth of L. lactis derivatives in CDM-Cel. (A) Black circles, MG1363 carrying a Pcel promoter up-mutation (k = 0.25 h−1); light gray circles, MGΔcelBΔptcCΔllmg_0963 llmg_1239::IS905 (M-IS) (k = 0.24 h−1); dark gray inverted triangles, MGΔcelBΔptcCΔllmg_0963 llmg_1239::gfp (M-GFP) (k = 0.23 h−1); light gray triangles, MGΔcelBΔptcCΔllmg_0963 (k = 0.0039 h−1). Growth curves show means for 3 replicates. (B) Genetic cluster llmg_1240-45 containing genes with predicted functions in sugar utilization and the predicted transcriptional repressor gene llmg_1239, located upstream. transcr. reg, transcription regulator; P-ase, phosphatase. (C) llmg_1239-llmg_1240 intergenic region. Translation start sites (ATG) are indicated by arrows, −10 and −35 sequences are underlined, and predicted operator sequences to which LacI family regulators can bind are shown in boxes. Black box, CcpA; red box, GalR of E. coli (note that L. lactis MG1363 does not possess GalR). The prediction was made using http://genome2d.molgenrug.nl.