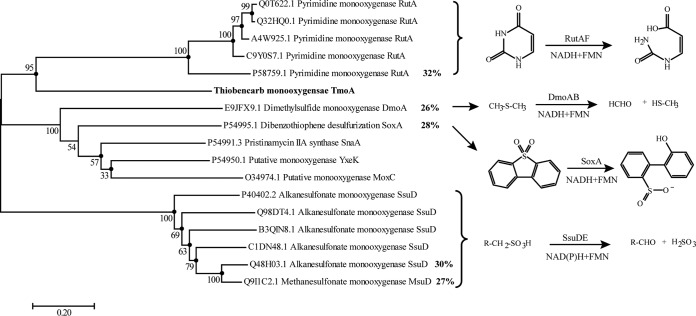
FIG 6.
Phylogenetic tree constructed based on the alignment of TmoA with the oxygenase components of certain related two-component flavin-dependent monooxygenases. Multiple-alignment analysis was performed with ClustalX v2.0, and the phylogenetic tree was constructed with the NJ, ML, and ME algorithms using MEGA 7.0; bootstrap values (based on 1,200 replications) are indicated at branch nodes. The solid circles indicate that the corresponding branches were also recovered using the ML and ME algorithms. Bar, 0.20 substitutions per nucleotide position. Each item was arranged in the following order: UniProtKB accession number and protein name.