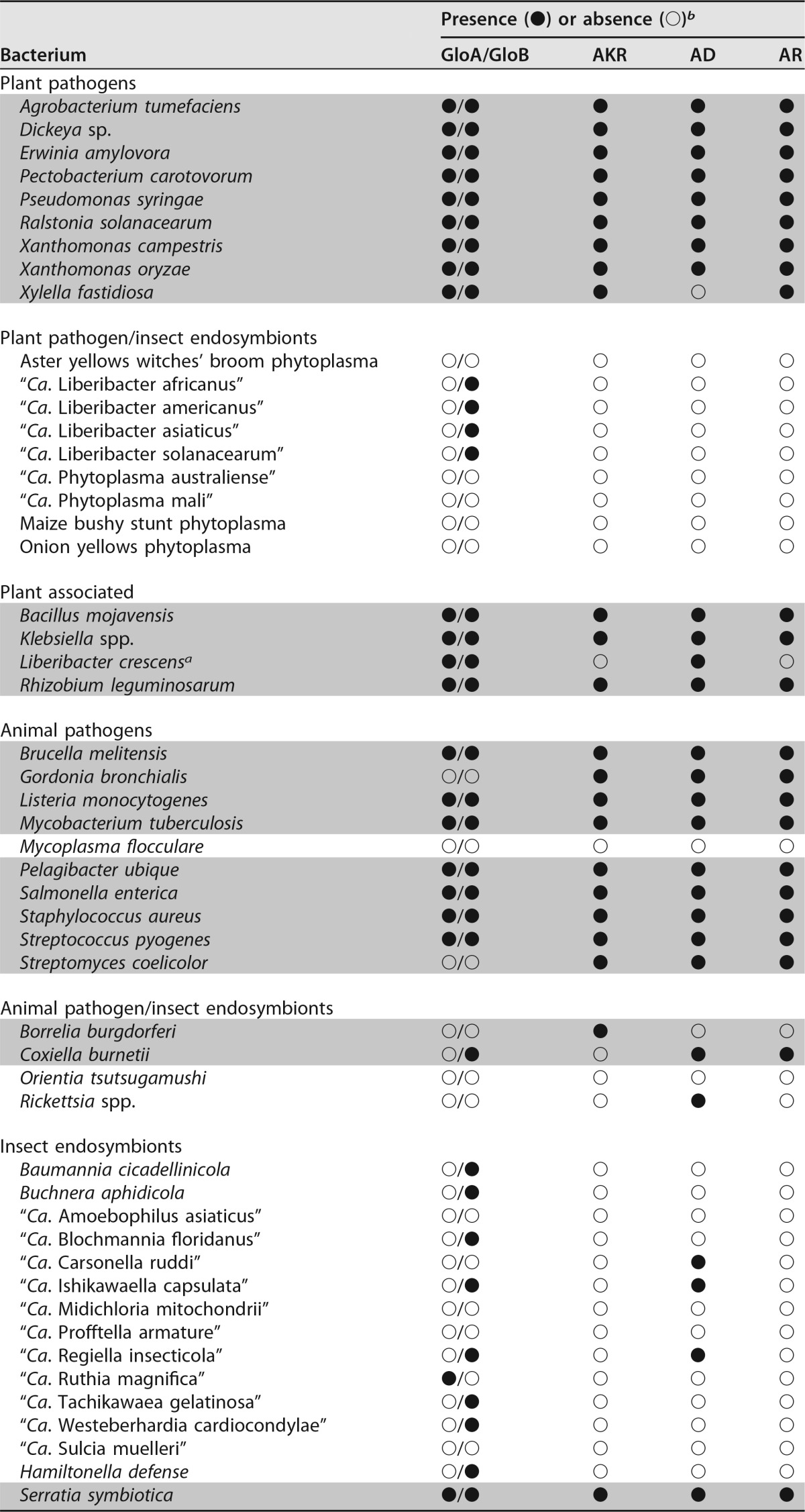
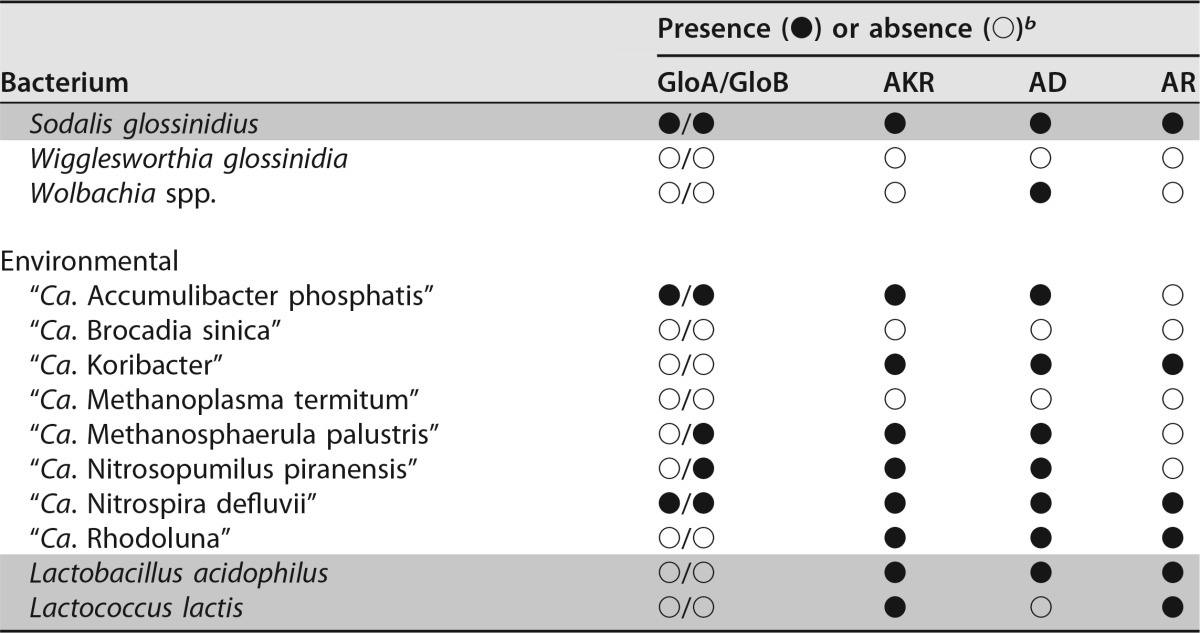
TABLE 1.
Survey of glyoxalase and nonglyoxalase methylglyoxal detoxification enzymes present in representative plant- and human-pathogenic, symbiotic, and environmental bacteria
a Plant and/or insect hosts are currently unknown for L. crescens.
b GloA, glyoxalase I (EC 4.4.1.5); GloB, glyoxalase II (EC 3.1.2.6); AKR, aldo/keto reductase superfamily (EC 1.1); AD, aldehyde dehydrogenase (EC 1.2.1.3); AR, aldehyde reductase (EC 1.1.1.2). The “●” and “○” symbols represent the presence or absence of either the glyoxalase or alternate (putative) methylglyoxal detoxification enzymes. They are marked as present in the subject species if any of the three query sequences used for each enzyme yielded values of >70% coverage with >30% identity. Gray-shaded rows denote culturability (defined as replicative growth) in axenic media.