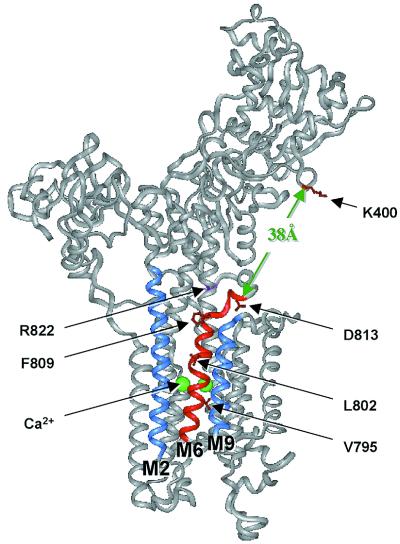
Figure 4.
Sites in the crystal structure of SERCA1a where PLN is proposed to interact. A version of the crystal structure of SERCA1a (2) was generated in the insight ii program (Molecular Simulations, Waltham, MA). PLN domain IA interacts with SERCA1a residues K397-V402 (red) in the nucleotide binding domain. PLN domain II binds to transmembrane helix M6 (red), lying in a groove between helices M2 and M9 (blue). Two Ca2+ ions (green) bind to the opposite face of M6. If PLN domain II is located in the groove formed by helices M2, M6, and M9 and if it is also associated with K400 in the cytosolic domain, then PLN domain IB must cross the region from the top of the M6 helix (G808) to D813 located in L67. This distance is about 10 Å. It must then cross a gap of about 38 Å to reach the cytosolic interaction site identified as K400. Evidence for interactions among residues N27 and N30 in PLN domain IB and residue D813 in SERCA1a L67 is fully consistent with this model for PLN interaction with SERCA1a. The crystal structure of SERCA1a was reprinted by permission from Nature (2), copyright 2000 Macmillan Magazines Ltd.