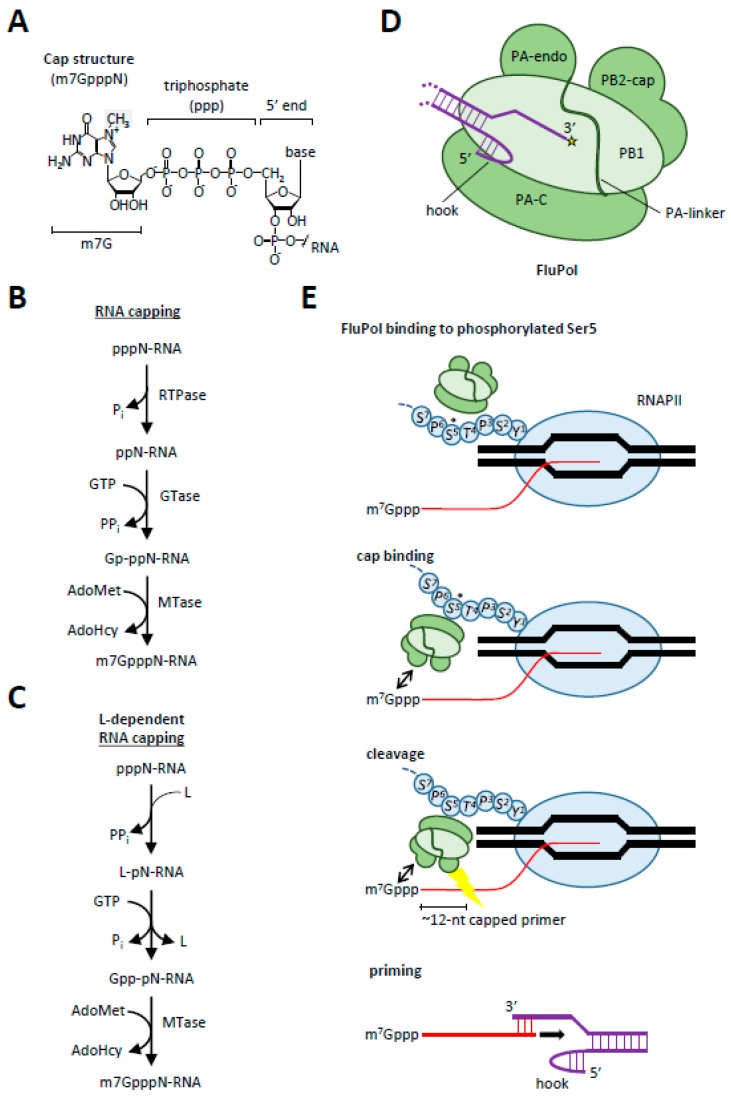
Figure 2.
Capping mechanisms in RNA viruses. (A) The eukaryotic mRNA cap consists of a 7-methylguanosine linked to the initiator nucleoside of mRNA through the 5′-5′ triphosphate bridge. The methyl group at the N7 position of the guanosine is shaded gray; (B) the conventional RNA capping pathway; (C) the L-dependent capping pathway utilized by negative-sense ssRNA viruses; (D) the FluPol complex with the polymerase acidic endonuclease (PA-endo), PA C-terminal (PA-C), polymerase basic 2 cap-binding (PB2-cap) and PB1 domains indicated. The influenza viral RNA (vRNA; purple) is circularized with the extreme 5′ end docked in a pocket formed by PA-C and PB1 and the 3′ end loaded in the FluPol active center (yellow star); (E) the first step in cap-snatching is the recognition and binding of FluPol to the phosphorylated Ser5 of RNAPII through its PA-C domain. This will allow the PB2-cap domain to bind the cap and to direct the nascent RNA towards PA-endo. After cleavage the nascent RNA will be loaded into the active domain of FluPol where it will be used to prime the 3′ end of the vRNA, allowing elongation of the RNA primer from 5′ to 3′ direction (indicated with black arrow).